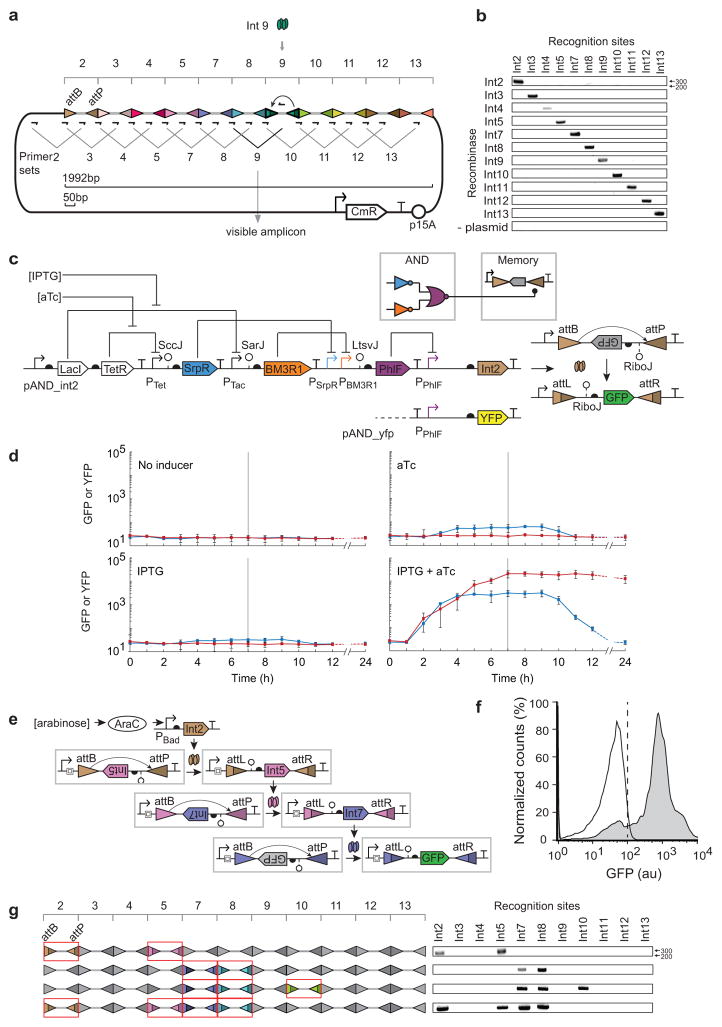
Figure 3. Incorporation of recombinases into larger genetic circuits. (a).
The memory array was designed as a linear concatenation of recognition sites for each integrase. A different 50bp spacer (grey thick line) was placed between each pair of recognition sites. Primer pairs were designed where one occurs at the interface between recognition sites and the second is within the spacer. Only when a spacer is inverted does the associated primer occur in the correct orientation to be amplified by PCR (~300bp). Int9 is shown as an example. (b) A DNA gel (1% agarose) is shown where the each primer set is used to determine which inversion event occurred. The “plasmid” control refers to a strain that contains only the memory array plasmid, but no integrase plasmid. (c) The memory array recording multiple bits of information is shown. Multiple integrase genes (Int2/5, Int7/8, Int7/8/10 or Int2/5/7/8) were organized in an operon controlled by an arabinose inducible promoter. The memory array after inversion is shown on the left. Only the attB/P sites that are switched are colored (same as part a) and indicated by the formation of attL/R. The DNA bands amplified using 11 primer sets described in Figure 3a are shown on the right. (d)The wiring diagram (colored by repressor) and genetic system for the AND gate connected to a memory switch is shown. The AND gate is also connected to an yfp gene to be used as a control. (e) Each panel shows a different combination of inducer. Blue lines show the fluorescence when the PPhlF promoter is fused directly to yfp and the red lines show when it induces Int2. Inducers were added at t = 0 hours and removed at t = 6 hours (horizontal line). The last time point was taken at 24 hours; the dashed lines are an extrapolation to that point accounting for dilution due to cell division. The error bars represent the standard deviation of three independent experiments performed on different days. (f) The 3-layer cascade of phage integrases is shown. Each integrase changes the orientation of the same constitutive promoter (BBa_J23101) and the same spacer and RiboJ insulator is used to insulate the integrases at each stage. (g) The cytometry distributions are shown for the cascade in the absence (white) and presence (grey) of inducer. The vertical dashed line demarcates the threshold used to determine whether cells are on or off. The figure represents three experiments performed in different days.