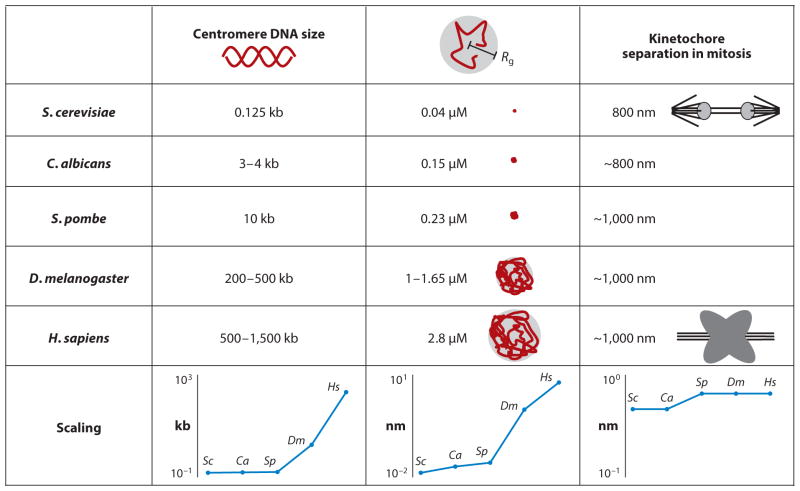
Figure 1.
Physical properties of centromere DNA. Centromere DNA size is defined as the region of DNA required for the segregation function in a variety of organisms. Rg is the radius of the random coil (radius of gyration) defined by Rg2 = n × (2Lp)2/6. Lp is the persistence length of the polymer, and n is the number of segments (total DNA contour length Lc/Lp) (61). Kinetochore separation is the distance between sister kinetochores observed experimentally in a number of organisms [budding yeast, S. cerevisiae (118); fission yeast, S. pombe (32); worm, C. albicans (94); Drosophila melanogaster (160); and human, H. sapiens (132)]. The bottom graphs highlight the scaling (ordinate) of DNA (four orders of magnitude), radius of gyration (three orders of magnitude), and kinetochore separation (constant) throughout phylogeny (abscissa).