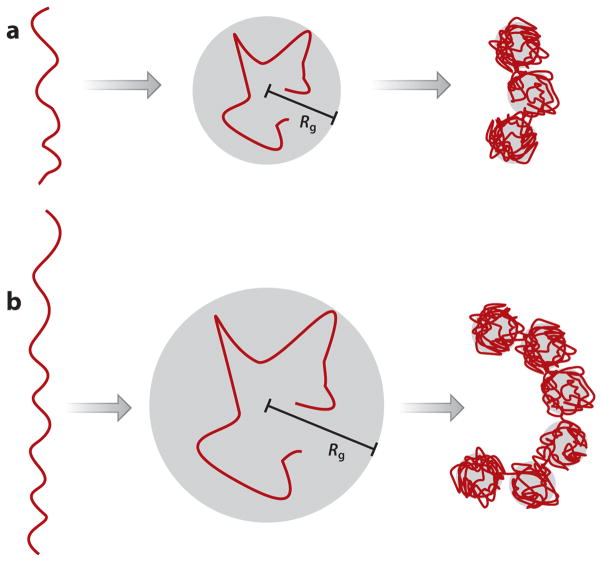
Figure 4.
Polymer model of chromatin provides insight into force transduction through the centromere heterochromatin. (a,b) Linear DNA (left) spontaneously folds into a random coil (middle). The size of the random coil is defined by the radius of gyration (Rg). In a confined and crowded environment such as a cell, the chains adopt smaller random coils within which monomer subunits are free to fluctuate, thus maximizing entropy (right, smaller gray spheres containing red polymer). These are known as thermal blobs and are defined as regions of the polymer chain in which the monomers are unaffected by their environment. If force is exerted at the ends of the chain, the blobs decrease laterally in size, but within each blob, the chains continue to fluctuate with defined statistical properties (164). Tension blobs create significant consequences for the centromere. When force is exerted on the kinetochore, tension can be transmitted over large distances (800 nm) without reducing the entropy of each and every monomer within the chain. The differing lengths of centromere DNA across phylogeny (small to large, as depicted in panels a and b) predict that the number of tension blobs varies considerably, but the mechanism of force transduction is highly conserved. The centroids of blobs can be connected to each other through springs, leading to the bead-spring representation of the chromatin polymer.