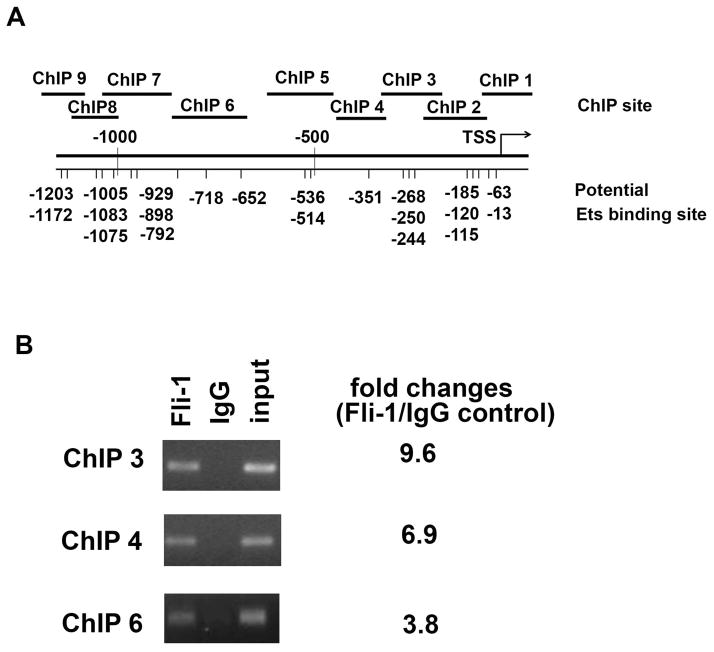
Fig. 5. ChIP analysis of Fli-1 binding to the IL-6 promoter.
The putative Fli-1 binding sites in the promoter region of the mouse IL-6 gene and the location of the primers used in the ChIP assay are shown in (A). MS1 endothelial cells were cross-linked with formaldehyde, chromatin was isolated from the cells and immunoprecipitated with specific Fli-1 or control IgG antibodies. The genomic fragments associated with the immunoprecipitated DNA were amplified by RT-PCR using primers specifically designed to cover putative Ets binding sites (The sequences of primers are available upon request to the corresponding author). PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Input represents 1% total cross-linked, reversed chromatin before immunoprecipitation. Fold changes were calculated by RT-PCR, ChIP1, ChIP2, ChIP5, and ChIP 7–9 sites were not significantly enriched with a specific antibody against Fli-1 (less than 1.3 fold change, not shown in the figure) (B). Data are representative of five independent experiments.