Fig. 1.
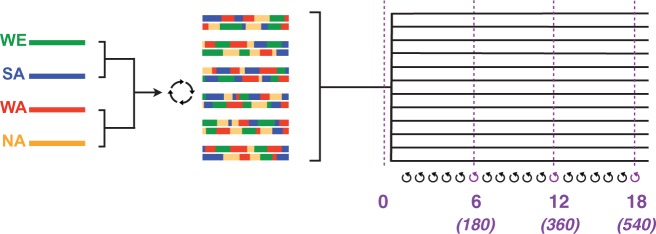
Schematic illustrating the experimental strategy. A four-way cross of diploid strains from different geographic origins (DBVPG6765, wine/European or WE in green; Y12, sake/Asian or SA in blue; DBVPG6044, W. African or WA in red; YPS128, N. American or NA in gold) generated a “synthetic population” that we used as the ancestral source for experimental populations (the SGRP-4X; Cubillos et al. 2013). Twelve replicate populations evolved in parallel and were sampled for sequencing at four timepoints: initially, and after 6, 12, and 18 rounds of forced outcrossing. Numbers in parentheses indicate the approximate number of mitotic divisions that have elapsed by each week.
