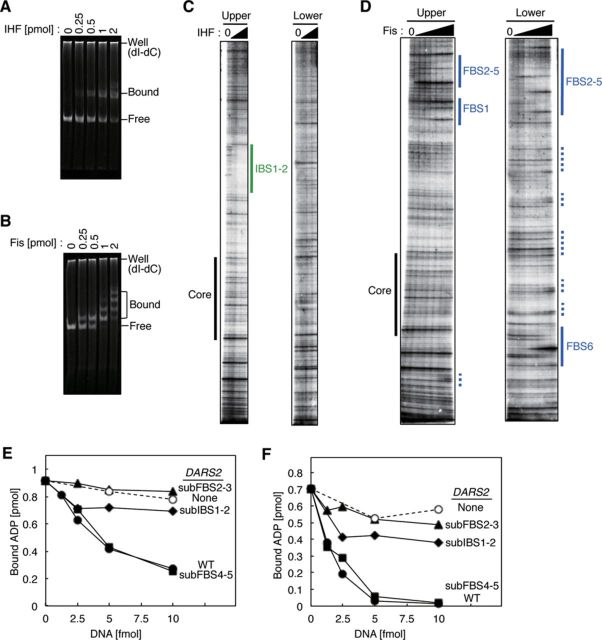
Figure 3.
DARS2 IBS1–2 and FBS2–3 are required for DARS2 activation in vitro. (A, B) Electrophoretic mobility shift assay. IHF (A) or Fis (B) was incubated for 5 min at 30°C in buffer GS containing DARS2 DNA (455 bp, 100 fmol) and poly (dI-dC) (120 ng), followed by 4% PAGE. Well; gel well, Bound; protein-bound DNA, Free; protein-free DNA. Poly (dI-dC) remained in the gel well. (C, D) Footprint analysis. The upper or lower strand of [32P]-DARS2 (100 fmol) was incubated at 30°C for 10 min with DNase I and IHF (0, 1.2, 2.4, or 4.8 pmol) (C) or Fis (0, 0.12, 0.24, 0.48, 0.96, 1.9, 3.8, or 7.6 pmol) (D), followed by urea–PAGE and phosphorimaging. Green lines: IBS1–2. Blue lines: FBS1 FBS2–5, and FBS6. Blue dotted lines: low affinity Fis-binding sites. For detailed sequences, see also Supplementary Figure S3. (E, F) Roles for DARS2 IBS and FBS in ADP dissociation from DnaA. MG1655 Fr II (5 μg) (E) or 50 fmol each of IHF and Fis (F) was co-incubated at 30°C for 15 min with [3H]ADP–DnaA (2 pmol) in the presence of pOA61 (WT) (•), pKX35 (subIBS1–2) (▴), pKX36 (subFBS2–3) (♦), pKX37 (subFBS4–5) (▪) or pACYC177 (None) (○). See also Supplementary Figures S4 and S5.