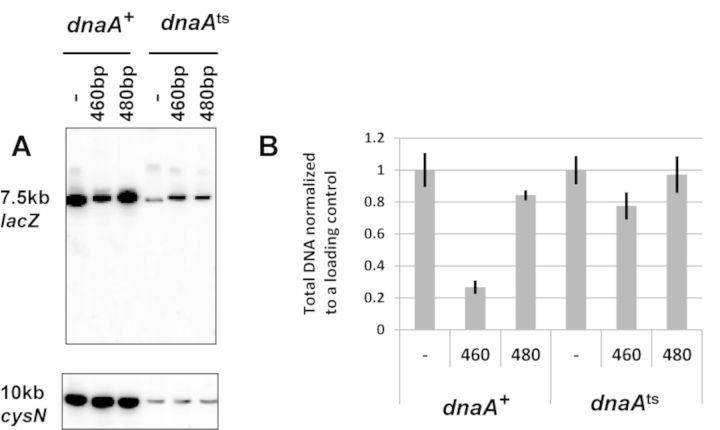
Figure 3.
Replication-dependence of DNA loss surrounding the break in recombination-proficient palindrome-containing cells. (A) Top panel. Southern blot showing the 7.5 kb NdeI lacZ fragment containing the palindrome. Lower panel. Southern blot showing the 10 kb NdeI cysN fragment (it can be seen that incubation at 42°C reduces the total yield of DNA). (B) Quantification of DNA loss. The total amount of DNA from each sample was quantified. DNA samples were taken after 60 min of growth in arabinose to induce expression of SbcCD. Strains used were DL2792 (lacZ+), DL3020 (lacZ::460bp) and DL3021 (lacZ::480bp), DL3584 (dnaAtslacZ+), DL3585 (dnaAtslacZ::460bp) and DL3586 (dnaAtslacZ::480bp). Error bars represent the standard error of the mean of three biological replicates. The difference in yields of DNA recovered between the dnaAts strains containing the 460-bp perfect palindrome and the 480 bp interrupted palindrome at 42°C was not statistically significant (unpaired two-tailed t-test p = 0.074).