Figure 4.
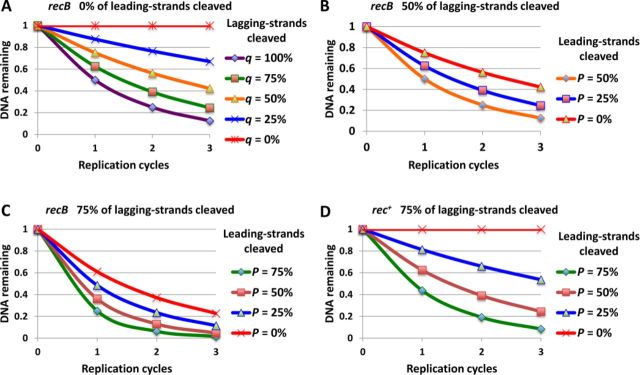
Predictions for DNA loss of alternative probabilities for cleaving leading- and lagging-strands during DNA replication. Expected DNA losses are presented for cleavage probabilities of 0, 25, 50, 75 and 100% as explained below. (A) Predicted DNA loss for cleavage of the lagging-strand in the absence of DSBR (recB mutant). This is the behaviour we observe for an interrupted palindrome (9) and the DNA loss we observe corresponds a frequency of strand-cleavage between 50 and 75%. We have therefore predicted (in B and C) the DNA loss obtained when both strands are cleaved using these two alternative probabilities of lagging-strand cleavage. (B and C) Predicted DNA loss for a 50% (B) and 75% (C) probabilities of lagging-strand cleavage coupled to probabilities of leading-strand cleavage of 0, 25, 50 and 75% in the absence of DSBR (recB mutant). In both cases we have assumed that the probability of leading-strand cleavage can be equal to that for lagging-strand cleavage but cannot exceed it. Our data are consistent with cleavage of both the leading- and lagging-strands of between 50 and 75%. (D) Predicted DNA loss for a 75% probability of lagging-strand cleavage coupled to probabilities of leading-strand cleavage of 0, 25, 50 and 75% in the presence of DSBR (rec+ cells). DNA is not expected to be lost in recombination proficient cells if the leading strand is not cleaved (0%). However, if it is cleaved, there will be no template for repair and DNA will be lost. Here, we have only illustrated the situation for 75% cleavage of the lagging-strand because the observed loss after three generations for the perfect palindrome corresponds to that predicted for cleavage of 75% of both strands.
