Figure 1.
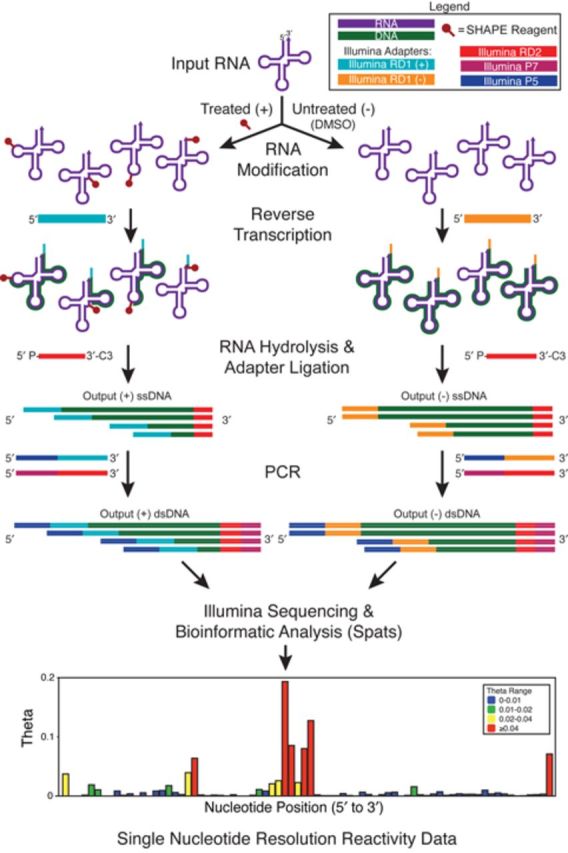
The basic SHAPE-Seq protocol. In SHAPE-Seq (6,7), RNAs are modified with chemical probes, such as 1M7 (24), or any probe that covalently modifies the RNA in a structure-dependent fashion (12). RT of the RNAs creates a pool of cDNAs, whose length distribution reflects the distribution of modification positions. Control reactions are performed to account for RT fall-off at unmodified positions. RT primer tails contain a portion of one of the required Illumina sequencing adapters, while the other is added to the 3′ end of each cDNA through a single-stranded DNA ligation. A limited number of PCR cycles are used to both amplify the library and add the rest of the required adapters prior to sequencing. A freely available bioinformatic pipeline Spats (7,26–27) is then used to align sequencing reads, correct for biases due to RT-based signal decay (26,27) and calculate reactivity spectra for each RNA. See Supplementary Figures S2–S4 for protocol details.
