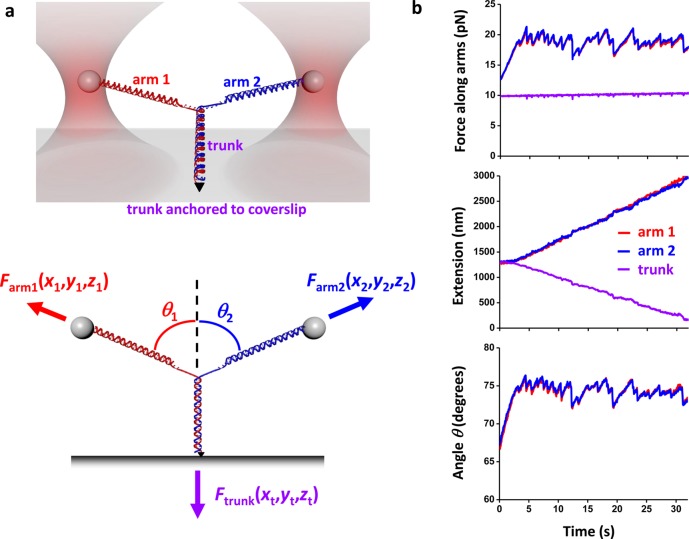
Figure 1.
Unzipping a Y structure. (a) Experimental configuration. (Top) The initial Y structure consisted of two dsDNA arms which were joined to a dsDNA trunk: one arm was attached to an optically trapped microsphere via a streptavidin/biotin connection; the second arm to a second optically trapped microsphere via a digoxigenin/antidigoxigenin connection; the trunk to a microscope coverslip via a fluorescein/antifluorescein connection. This version of the Y-structure contained a single anchoring point of the trunk via one of its two DNA strands and thus permitted the trunk end to swivel around the anchoring point without any torsional constraint. (Bottom) Y structure geometry and force balance. The trunk dsDNA was mechanically unzipped by pulling on the arms with the two optical traps. The force vector on each arm was independently measured, and thus the force on the trunk was determined by force balance at the junction. The 3D position of each trapped microsphere and the trunk anchoring point were also measured. (b) An example data trace from symmetric unzipping of the trunk of a Y structure under a constant force on the trunk. The force and extension of each branch of the Y-structure as well as the angles of the Y-structure were measured as functions of time. Data were taken at 10 kHz and filtered to 20 Hz.