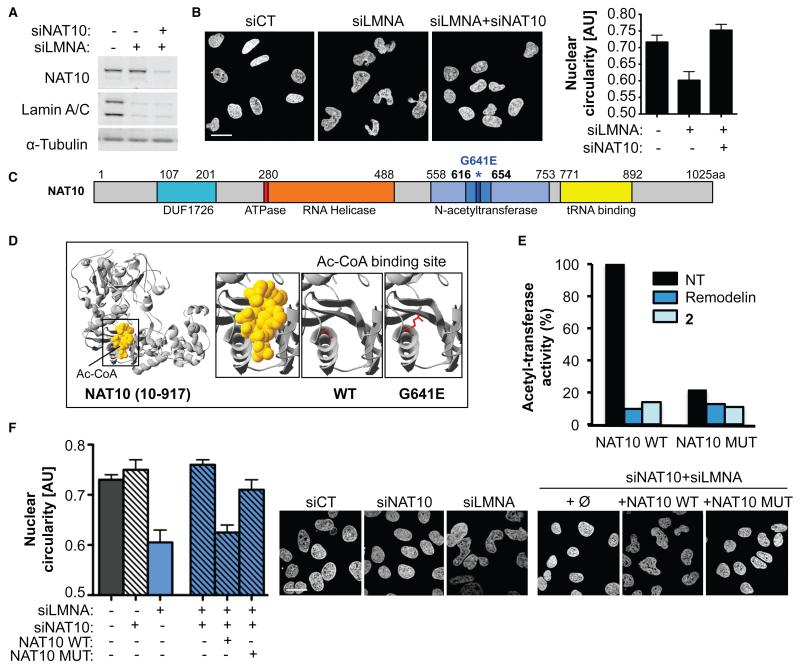
Figure 2. Inhibiting NAT10 activity by Remodelin mediates nuclear shape rescue of LMNA depleted cells.
A) NAT10 and Lamin A/C depletion in U2OS cells. B) Nuclear shape visualized by DAPI staining (left) and quantification of nuclear circularity (right; means of three independent experiments with n>267 ± s.d.). Scale bar: 20 μm. C) Representation of NAT10 with its known domains. The G641E mutation identified in D) is indicated in dark blue and asterisked. D) Modelled 3D structure of human NAT10 residues 10-917 showing the acetyl-CoA binding site (left) and disruption of Ac-CoA binding by NAT10 G641E mutation (right) visualized with Swiss-Prot PDB Viewer. E) In vitro acetylation assay showing the activity of NAT10 towards tubulin. F) Quantification of nuclear circularity (left) in cells stably expressing siRNA-resistant FLAG-NAT10 WT or FLAG-NAT10 G641E (NAT10 MUT; means of three independent experiments with n>198 ± s.d.) and nuclear shape visualized by DAPI staining (right). Scale bar: 20 μm.