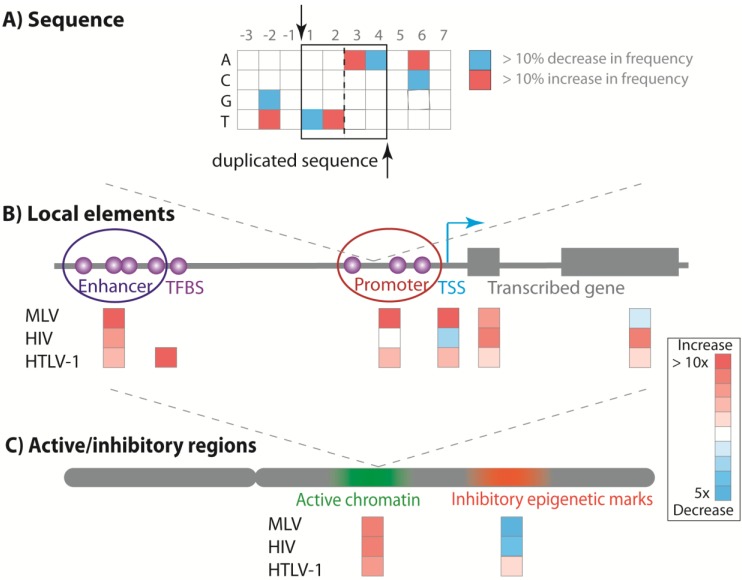
Figure 1.
Genomic features associated with retroviral integration sites. The site of retroviral integration is determined by factors at several physical scales. (A) Each viral integrase has a weak preference for a short characteristic genomic sequence (the MLV preference is illustrated here, based on data in [44]); (B) Host proteins interact with the viral integrase and guide integration into genes and regulatory elements, such as promoters and enhancers, and near to (100–1000 bp) transcription start sites (TSS) and transcription factor binding sites (TFBS); (C) The frequency of integration differs between regions of the genome marked respectively by inhibitory and activatory epigenetic marks. The heat map marks a typical fold change in integration frequency compared with random sites, based on data in [39,49,55].