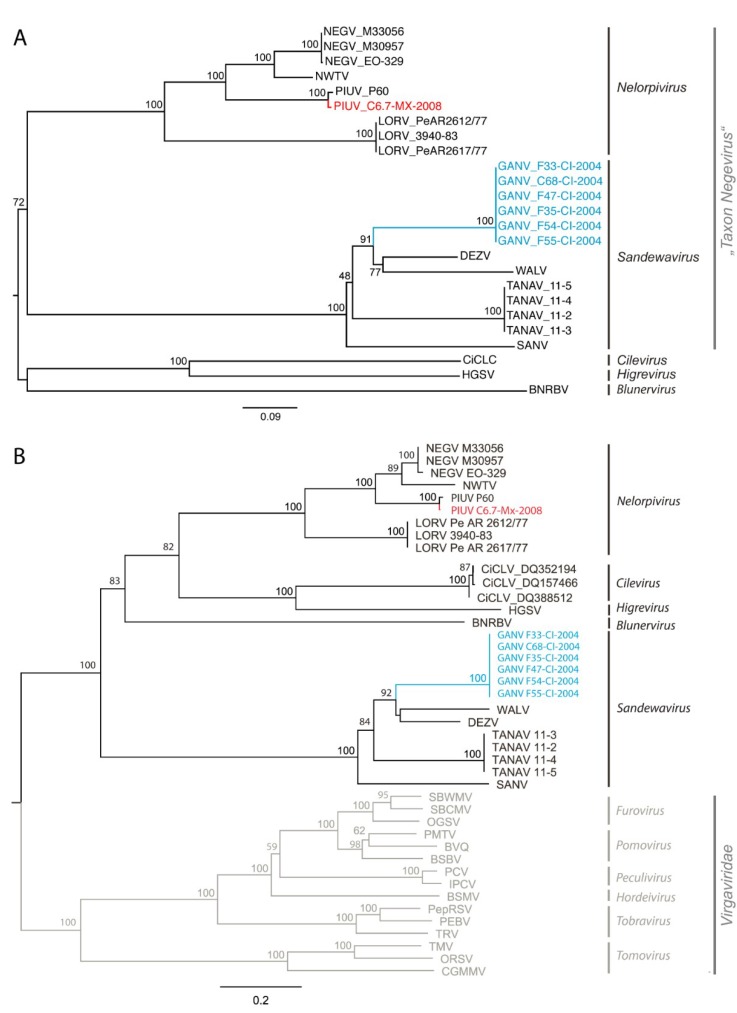
Figure 1.
Phylogenetic relationship of Goutanap virus and Piura virus C6.7-MX-2008. (A) Analysis of the conserved domains of the RNA-dependent RNA polymerase proteins of negeviruses, cileviruses, higreviruses and blunerviruses; (B) Analysis of a gap-free concatenated alignment of fused methyltransferase, helicase and RNA-dependent RNA polymerase domains of members of negeviruses, cileviruses, higreviruses and blunerviruses, as well as representative members of each genus of the Virgaviridae family. ML analyses were based on an 805 amino acid alignment (A) and a 445 amino acid alignment (B) guided by the Blosum62 (A) and Dayhoff (B) substitution matrix using PHYML as implemented in Geneious. Confidence testing was performed with 1000 bootstrap replicates. Virus names and GenBank accession numbers are provided in Table S1.