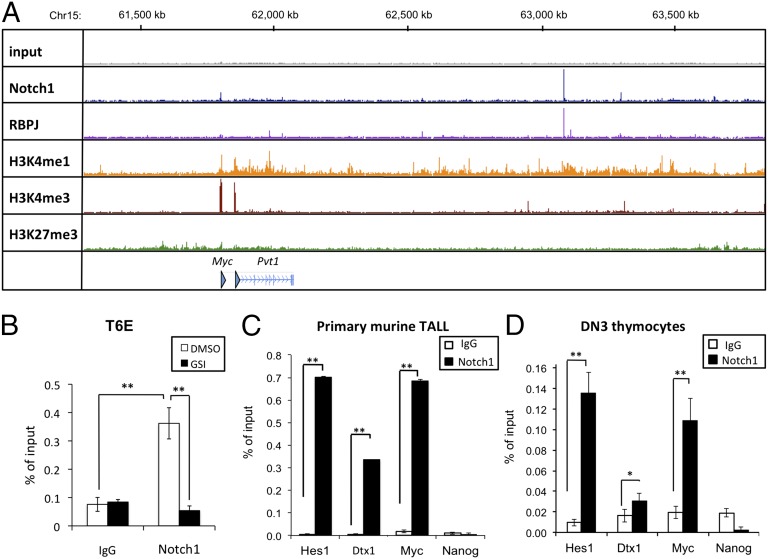
Fig. 1.
Identification of a site ∼1.27 Mb 3′ of the Myc gene body that binds RBPJ and Notch1 in normal and transformed murine pre-T cells. (A) ChIP-Seq read counts for Notch1, RBPJ, and H3K4me1, H3K4me3, and H3K27me3 marks in a ∼2-Mb region containing Myc. y axis, aligned reads; arrowhead, TSSs. (B) Local ChIP for Notch1 at the 3′ Myc site (NDME) in the murine T-ALL cell line T6E treated with GSI or DMSO for 4 h before harvest. Nonspecific Ig was used as a control. (C and D) Local ChIP analyses of Notch1 occupancy of the binding site 3′ of Myc (NDME) in primary murine T-ALL cells (C) and DN3 thymocytes (D). Previously characterized Notch1 binding sites in the Hes1 promoter and Dtx1 intron2 serve as positive controls; Nanog serves as a negative control. In B–D, data were obtained in triplicate in independent experiments; error bars correspond to the SEM. *P < 0.01; **P < 0.001.