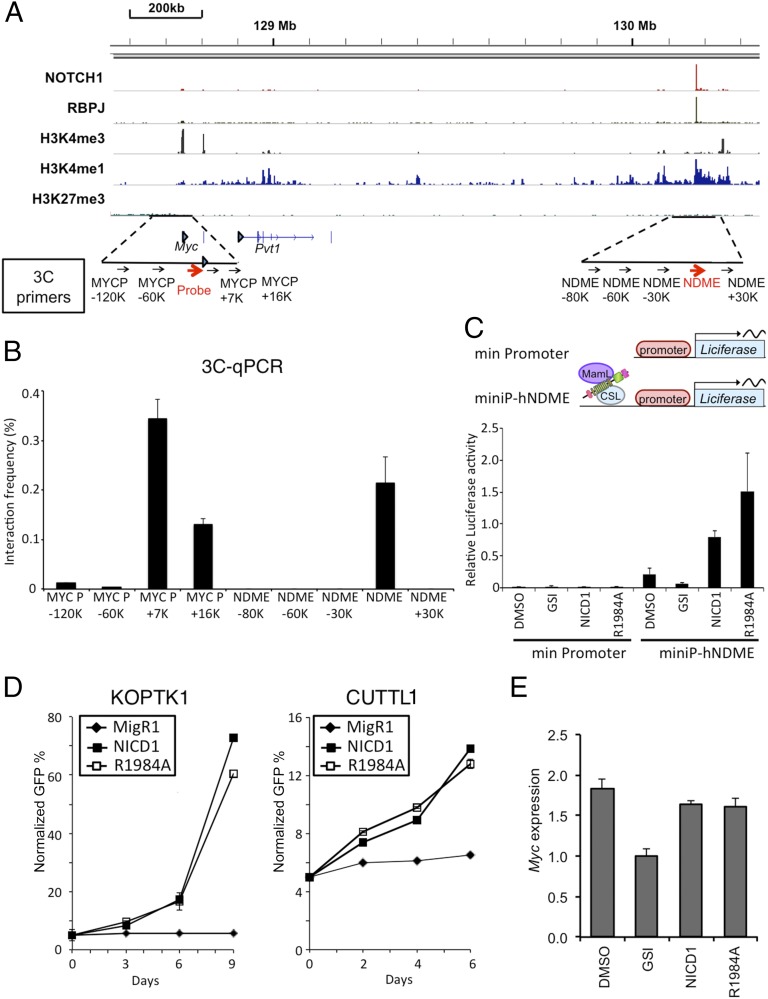
Fig. 4.
Notch1 regulates human Myc via a 3′ enhancer element. (A) ChIP-Seq read counts for Notch1, RBPJ, and H3K4me1, H3K4me3, and H3K27me3 marks in a ∼2-Mb region containing human Myc. y axis, aligned sequence tags; arrowhead, TSSs. (B) The 3C assay in the human T-ALL cell line CUTLL1. Graphs show the enrichment of PCR product normalized to HindIII-digested, randomly ligated BAC DNA spanning the region. The arrows beneath the schematic in A denote the position of primers used for 3C analysis. The Myc promoter primer and “probe” were paired with eight primers positioned in flanking sequences to detect ligated HindIII-digestion fragments. (C) The human Myc 3′ enhancer element (NDME) stimulates transcription. (Upper) Schematic of the human Myc NDME and control firefly luciferase reporter genes. A minimal TATA promoter construct was used as a negative control. Note: RBPJ is denoted by its alternative name, CSL. (Lower) CUTLL1 cells were transfected with the indicated reporter genes with and without 1 µM compound E. Luciferase assays were performed 48 h later. (D) KOPT-K1 (Left) and CUTLL1 (Right) cells transduced with either NICD1, NICD1-R1984A, or empty plasmid were cultured in the presence of 1 μM compound E beginning 24 h after transduction. The number of GFP+ transduced cells was determined at various time points and normalized to time 0 (defined as 48 h after transduction). (E) Myc mRNA expression in CUTLL1 cells transduced as in D. Except for the MigR1 empty vector control cells, 1 μM compound E was added to cells 24 h after transduction. Cells were harvested for RT-PCR analysis 48 h later. In B–E, data were obtained in triplicate in independent experiments; error bars correspond to the SEM.