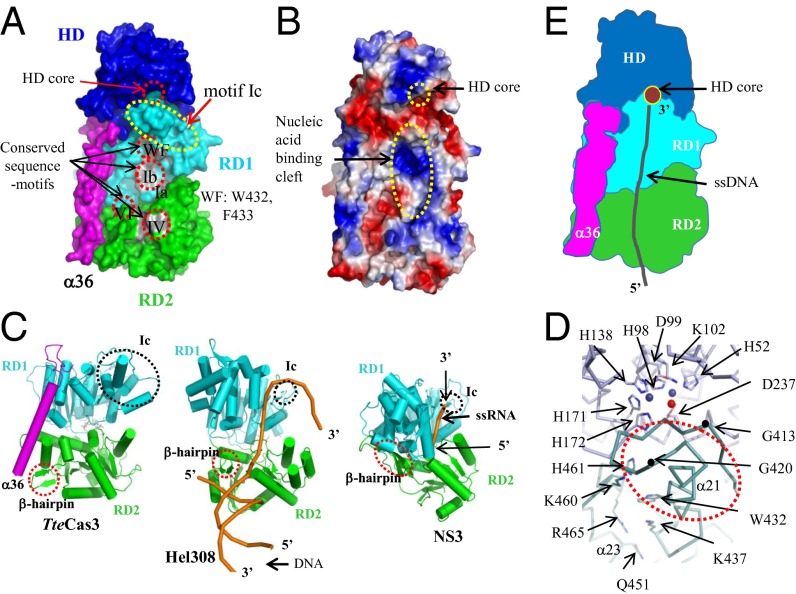
Fig. 3.
Disassembly line formed by aligned HD nuclease and helicase motifs of TteCas3. (A) Surface presentation of TteCas3. CTD has been omitted to show conserved regions on RD1 and RD2. Each segment is discerned by alternating colors. Conserved sequence motifs are labeled and highlighted by gray patches and dotted circles. (B) Surface potential map. The surface potential formed on NTD, RD1, α36, and RD2 is shown. The HD core and nucleic acid-binding cleft on both RDs were indicated. (C) Comparison of RDs from three closely related SF2 helicases. The structures of TteCas3 (Left) and Hel308 (Center) and NS3 (Right) helicases are oriented similarly. β-hairpin and motif Ic are indicated with circles. The bound nucleic acids on the Hel308 and NS3 helicases are drawn with orange ribbons. (D) The interface of HD (blue) and RD1 (cyan). A part of the TteCas3 extended Ic-motif helix is indicated. Conserved residues are displayed with stick models, and two Mn ions and a water molecule at the HD core are displayed with alternating balls. One motif Ic loop is highlighted by black dots representing residues Gly413 and Gly420. (E) Schematic diagram to show the ssDNA path to the HD core from the helicase fragment. Each domain is differentiated by colors, and the ssDNA is displayed by a black coil.