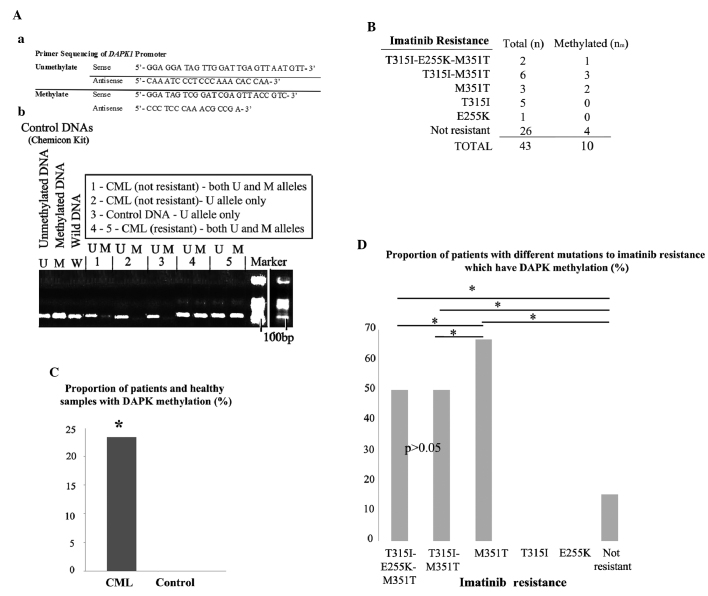
Figure 1.
Analysis of DAPK1 methylation. (Aa) DAPK primer sequences used for methylation analysis performed using methylation-specific PCR following bisulfite conversion. (Ab) Representative PCR products for U and M alleles of DAPK1 in resistant (samples 4 and 5) and non-resistant (samples 1 and 2) patients, in a control sample from healthy individuals (sample 3), as well as in control U, M and W DNA samples. (B) The total number of samples and the number of samples exhibiting DAPK1 methylation. (C) The proportion (%) of samples with DAPK1 methylation is significantly greater in CML patients compared with the healthy controls. (D) The proportion (%) of patients with DAPK1 methylation varied among different mutations. The majority of patients with M351T demonstrated DAPK1 methylation; however, none of patients with T315I or E255K were methylated, and no difference was detected between the patients with triple (T315I-M351T-E255K) and double (T315I-M351T) mutations (P>0.05). Furthermore, a relatively low number of non-resistant patients demonstrated DAPK1 methylation (P<0.05). *P<0.05. DAPK1, death-associated protein kinase-1; PCR, polymerase chain reaction; U, unmethylated; M, methylated; W, wild type; CML, chronic myeloid leukemia.