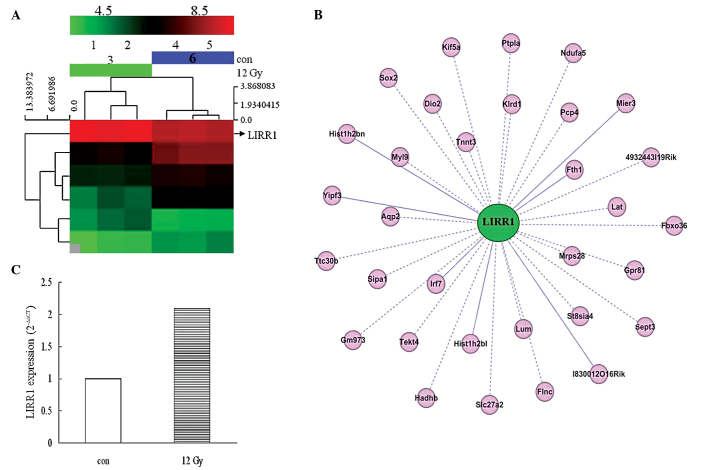
Figure 1.
Screening, validation and analysis of differentially-expressed large intergenic non-coding RNA-LIRR1 in lung tissue with acute radiation-induced lung injury. (A) Hierarchical cluster analysis diagram of 12-Gy X-ray irradiated lung tissues (n=3) vs. normal lungs (n=3). The arrow pointing to the LIRR1 gene and the color saturation varying from green to red represent the log2 value of the normalized intensity of each sample, ranging between 4.5 and 8.5 (P-value limit, 0.05). The rows indicate the individual probe sets, and the columns denote the experimental sample. (B) Co-expression network of LIRR1 and the encoding genes. The R-value was calculated to define the correlation coefficient, PCC, between LIRR1 and the encoding genes. A value of ≥0.99 was considered to indicate a co-expression pair. CytoScape software was used to integrate the interaction network between LIRR1 and the encoding genes. The pink nodes indicate the encoding genes and the green node denotes LIRR1. A solid line between two nodes indicates a positive correlation and a dotted line indicates a negative correlation. (C) LIRR1 transcription was validated in normal mouse lung tissues (white bar) and lung tissues from mice exposed to total body irradiation for 24 h (striped column; n=3 for each treatment). Reverse-transcription polymerase chain reaction was conducted in triplicate for each individual sample. All the experiments were performed independently at least three times. LIRR, long intergenic radiation-responsive non-coding RNA; con, control.