Figure 2.
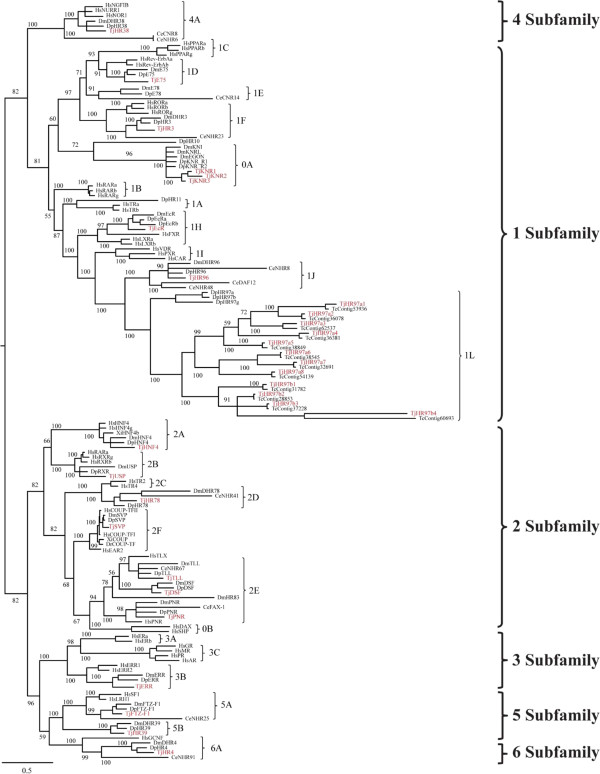
Gene phylogeny of nuclear receptors in T. japonicus and other related species. Phylogenetic distance was calculated with combined DBD-LBD amino acid sequences from T. japonicus and other species. A best-fit substitution model was established using maximum likelihood (ML) analysis supported by MEGA (ver.6.0). Numbers at nodes represent ML bootstrap support values and Bayesian posterior probabilities (=1.00). Details of model testing and parameters are provided in the Methods section. Tree is proportionally scaled, with the scale bar indicating sequence distance as number of substitutions. Species abbreviations: Ce: Caenorhabditis elegans, Dm: Drosophila melanogaster, Dp: Daphnia pulex, Dr: Danio rerio, Gg: Gallus gallus, Hs: Homo sapiens, Tj: Tigriopus japonicus, Xl: Xenopus laevis.
