Abstract
G-quadruplexes (four-stranded DNA secondary structures) are showing promise as new targets for anticancer therapies. Specifically, G-quadruplexes in the proximal promoter region of regulatory genes have the potential to act as silencer elements and thereby turn off transcription. Thus, compounds that are capable of binding to and stabilizing G-quadruplexes would be of great benefit. In this chapter we describe two recent studies from our labs. In the first case, we use NMR to elucidate the structure of a 2:1 complex between a small molecule and the G-quadruplex in the c-MYC promoter. In the second case, we use an allele-specific transcription assay to demonstrate that the effect of a G-quadruplex-interactive compound is mediated directly through the G-quadruplex. Finally, we use this information to propose models for the interaction of various small molecules with the c-MYC G-quadruplex.
Keywords: G-quadruplex, c-MYC, gene regulation, NMR structure
1 Introduction
The initial target for anticancer drugs was DNA and, today, drugs that target DNA remain the mainstay of most chemotherapy regimens [1]. However, interest in developing DNA-targeting drugs has waned due to a lack of selectivity for a particular sequence or region. Additionally, new and more specific molecular targets such as kinases and cell surface receptors have been identified for cancer. Recently, the discovery that telomeres and some guanine-rich (G-rich) promoter regions can form four-stranded DNA secondary structures termed G-quadruplexes [2–6] has ushered in an opportunity for a new phase of more selective DNA-targeted therapeutics.
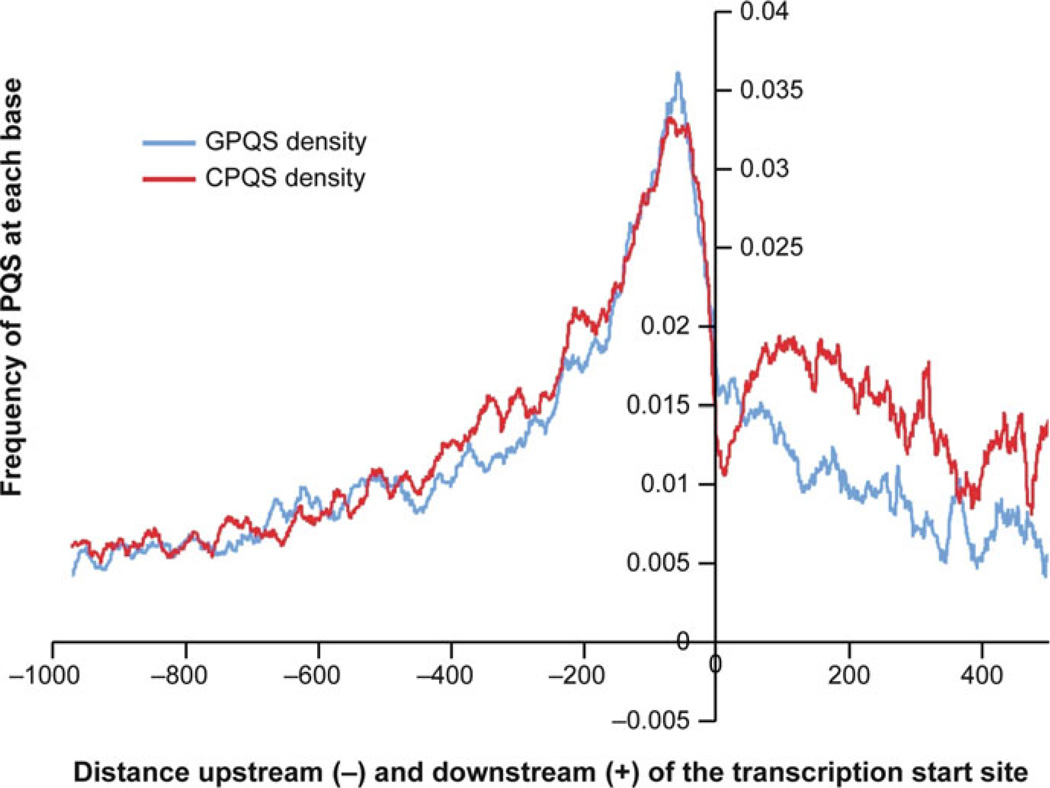
The ability of G-rich telomeres to form higher order DNA structures was described in 1988 [4]. Telomeres occur at the end of eukaryotic chromosomes and contain large numbers of simple guanine-rich tandem repeats. The enzyme telomerase contributes to cell immortalization by catalyzing telomere extension and is over-expressed in a number of human cancers. In 1997 telomerase was shown to be inhibited by ligands that interact with G-quadruplexes in the 3′-end of human telomeric DNA [7]. Since then, bioinformatics studies have been performed to investigate the incidence of putative G-quadruplex-forming sequences (GPQSs) throughout the genome [3, 8–10]. These studies revealed GPQSs to be enriched in the proximal promoter regions of regulatory genes, especially of proto-oncogenes, in an evolutionarily conserved manner [2, 3]. Figure 1 shows the frequency (or probability) of each nucleotide upstream or downstream of the transcriptional start site being part of a GPQS [10, 11]. This frequency peaks ~50 nucleotides upstream of the transcriptional start site, a region known for transcription regulation [10–12]. This clustering of GPQSs in known gene regulatory regions suggests that they may play a role in transcriptional control.
Fig. 1.
Frequency plot showing the probability of each nucleotide upstream (−) or downstream (+) of the transcriptional start site being part of a putative G-quadruplex-forming sequence (GPQS), based on the Quadparser algorithm [9]. The data have been averaged over all human protein coding genes in the genome. The blue plot represents the presence of a G-quadruplex motif (GPQS) while the red plot represents the C-rich complement of a G-quadruplex motif (CPQS). Figure is modified, with permission, from [11], © (2008) Oxford University Press
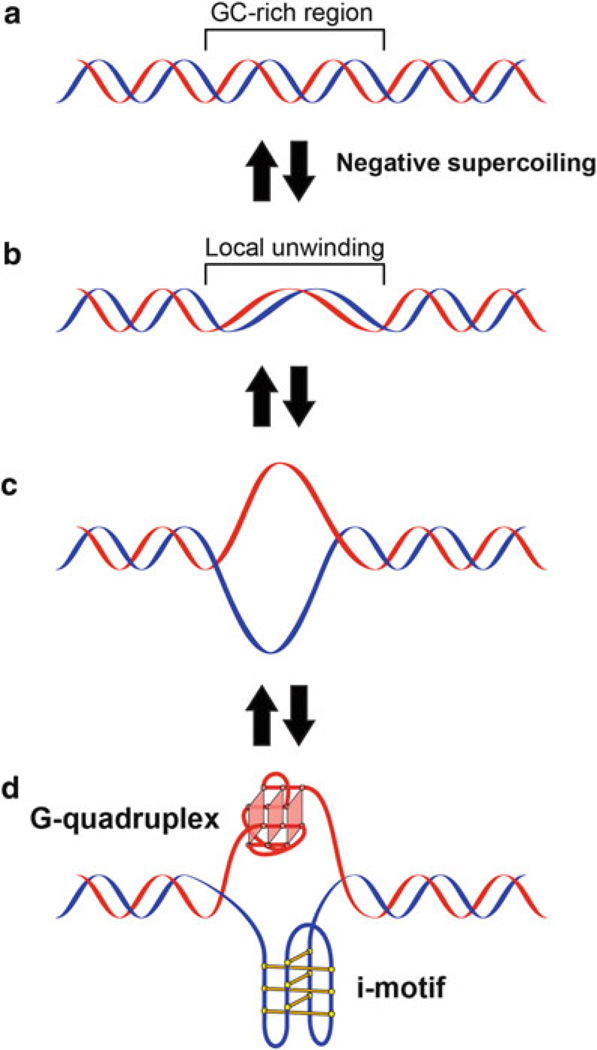
DNA supercoiling, a major source of superhelical stress in cells, is also known to play an important role in transcription [13]. As the transcriptional machinery translocates down a gene, it generates positive supercoiling downstream and negative supercoiling upstream. This negative supercoiling results in local unwinding of DNA and facilitates the opening of the guanine-rich/cytosine-rich (GC-rich) regions of the DNA. This process is thought to provide the equilibrating energy that facilitates the transition from double-stranded DNA to higher order non-B-DNA structures, allowing for the formation of G-quadruplexes on the G-rich strand and a complementary DNA secondary structure (i-motif) on the C-rich strand (Fig. 2) [15].
Fig. 2.
Proposed equilibrating forms of DNA produced under negative supercoiling induced by transcription: (a) duplex, (b) locally unwound, (c) single stranded, (d) G-quadruplex/i-motif structure. Figure is modified, with permission, from [14], © (2009) American Chemical Society
G-quadruplexes exhibit a diverse range of folding patterns: they are classified primarily by their tetrad directionality, loop length, and constitution [15]. These complex structures are highly polymorphic and capable of exhibiting parallel, antiparallel, or mixed topologies. The greatest sources of variability in G-quadruplexes are the lengths of their loops, which commonly range from one to nine base pairs, and their base composition, which has no constraints. On occasion, the loops themselves have been noted to form alternate DNA structures. For example, the G-quadruplex in the hTERT promoter contains a 26-base-pair loop observed to form a hairpin structure [15]. We have recently demonstrated that this tertiary DNA structure shows cooperativity in folding and unfolding such that a higher order interaction takes place between the hairpin and the G-quadruplex. This is the first example of cooperative refolding in DNA, a process previously documented only in proteins and complex RNA structures [16]. The unique globular shape and folded structures of G-quadruplexes and their potential ability to regulate the transcription of a host of oncogenes make them an attractive drug target.
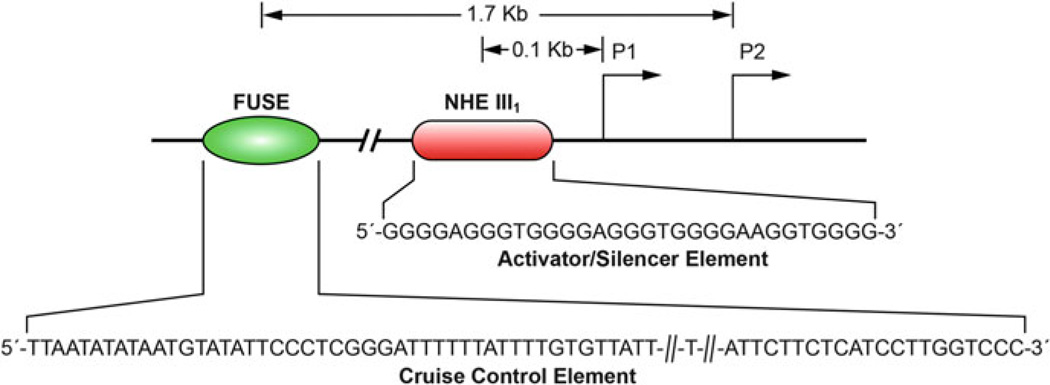
Perhaps the most extensively studied and well-characterized promoter G-quadruplex is within the promoter of the c-MYC proto-oncogene, which plays a role in normal cell proliferation and differentiation [17]. Transcriptional control of the c-MYC proto-oncogene has been investigated and described in numerous studies [2, 18, 19]. From these, two key regulatory regions have been identified: the far upstream element (FUSE) and the proximal nuclease hypersensitive element III1 (NHE III1) (Fig. 3). Using a mechanosensor mechanism fueled by transcriptionally induced negative supercoiling, the FUSE has been shown to function as a cruise control element [13]. The GC-rich NHE III1, upstream of the core promoter region, is responsible for 80–90% of c-MYC transcription [19]. It consists of five repeats of the sequence TGGGGA(G/A)G(G/A) and serves as an activation/silencing region for c-MYC transcription [20]. Under negative supercoiling conditions, this region has been shown to forma G-quadruplex, which acts as a silencer element [14, 17, 21, 22].
Fig. 3.
The position and sequence of the FUSE element and NHE III1 relative to the P1 and P2 promoters of c-MYC. The sequence shown for the FUSE element is only a partial representation. Figure is modified, with permission, from [18], © (2009) Macmillan Publishers Limited
c-MYC is aberrantly expressed, most often through an increase in transcription, in an estimated 80% of all human malignancies [17]. Thus, there is a considerable therapeutic value in the targeted down-regulation of c-MYC. In fact, Soucek and co-workers showed that systemic c-MYC inhibition in Ras-induced lung adenocarcinoma mouse models leads to regression of lung tumors [23]. Furthermore, while the systemic inhibition of c-MYC showed an effect on tissue regeneration, the effects were well tolerated and reversible [23]. With this in mind, we used NMR modeling and biological characterization to target the G-quadruplex in the c-MYC promoter in an attempt to find compounds that could down-regulate the expression of c-MYC at the transcriptional level.
2 NMR Determination of the Quindoline-i:c-MYC G-Quadruplex Complex
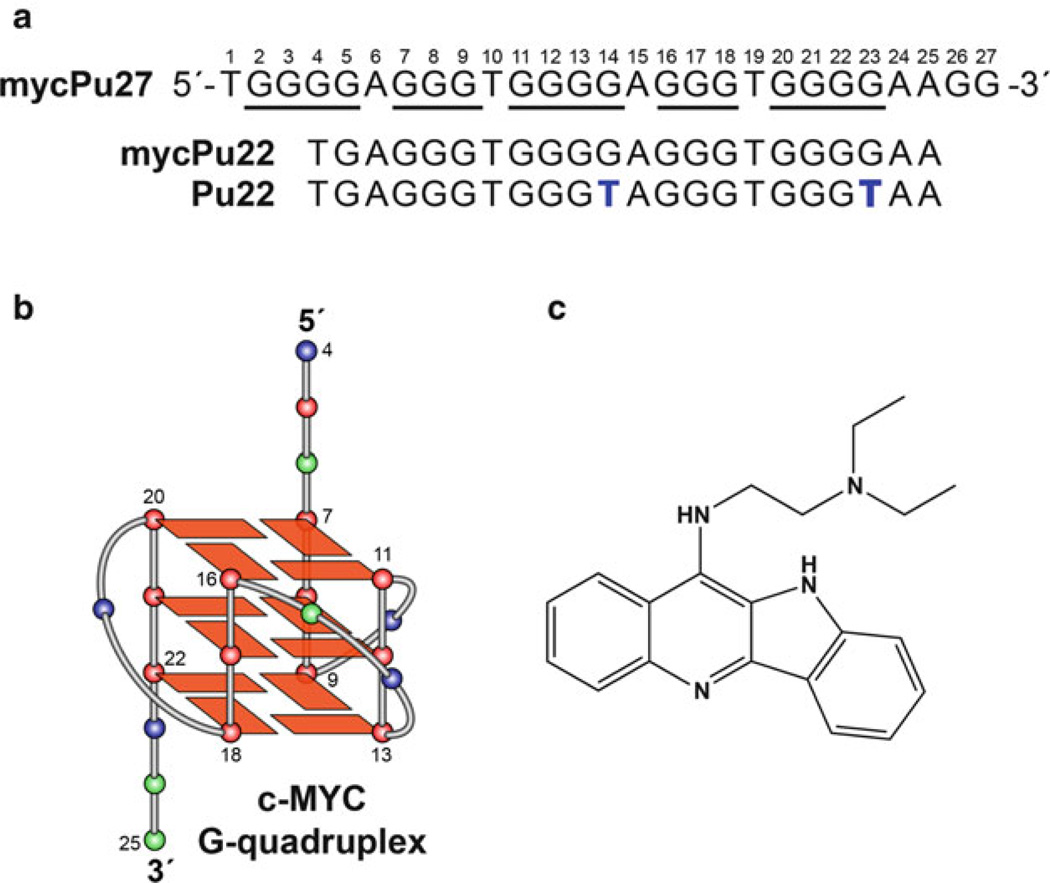
We previously determined the K+ solution structure of the major G-quadruplex formed in the c-MYC promoter. This c-MYC promoter is a parallel-stranded structure with two G3-N-G3 single-nucleotide double-chain-reversal loops, represented by the modified c-MYC promoter sequence, Pu22 (Fig. 4a, b) [25]. The G3-N-G3 motif has been shown to be a stable and prevalent structural motif in G-quadruplexes within a gene promoter [26]. Although the list of molecular structures reported for G-quadruplexes formed in gene promoters is growing, their structures with small molecules have been more difficult to obtain. We have very recently determined the NMR structure of a 2:1 complex of quindoline-i and the c-MYC G-quadruplex (PDB code: 2L7V) [24], which represents the first drug complex structure of a biologically relevant unimolecular promoter G-quadruplex.
Fig. 4.
(a) The promoter sequences of the NHE III1 of the c-MYC gene and its modifications. mycPu27 is the wild-type 27-mer G-rich sequence of the c-MYC NHE III1. mycPu22 is the wildtype 22-mer G-rich sequence of the c-MYC NHE III1 that forms the major G-quadruplex in physiologically relevant K+ solution. Pu22 is the modified mycPu22 sequence, with G-to-T substitutions at positions 14 and 23, that forms the predominant c-MYC promoter G-quadruplex in K+ solution and whose structure was determined by NMR. (b) The folding topology of the c-MYC G-quadruplex adopted by Pu22. Red box/red ball = guanine; green ball = adenine, blue ball = thymine. (c) The quindoline-i molecule. Figure is modified, with permission, from [24], © (2011) American Chemical Society
The quindoline-i compound (Fig. 4c) is a derivative of the natural product cryptolepine [27, 28]. This compound has been shown to stabilize the G-quadruplex formed in the c-MYC promoter and subsequently inhibit the expression of c-MYC in the hepatocellular carcinoma cell line H2p G2 [29]. The structure showed an unexpected drug-induced reorientation of the flanking sequences at both ends of the DNA sequence. This reorientation, called an “induced intercalated triad pocket,” acts as a mode of recognition and is analogous to the process used by riboswitches [30]. This mode of binding is distinct from previously proposed models wherein the planar G-quadruplex-interactive compounds such as telomestatin [31] and TMPyP4 [32] stack on the external guanine tetrad (G-tetrad). Additionally, the NMR structure of the quindoline-i:c-MYC G-quadruplex complex indicates that asymmetric compounds with a crescent shape, appropriate functional groups, and a small stacking moiety are more likely to bind in a defined manner to a unimolecular parallel-stranded G-quadruplex. This study describes the importance of the ligand shape as well as the two flanking bases of the G-quadruplex in determining drug-binding specificity and provides important insights for the structure-based rational design of molecules that interact with unimolecular parallel-stranded G-quadruplexes commonly found in promoter elements.
2.1 Solution Structure of the 2:1 Quindoline-i:c-MYC G-Quadruplex Complex
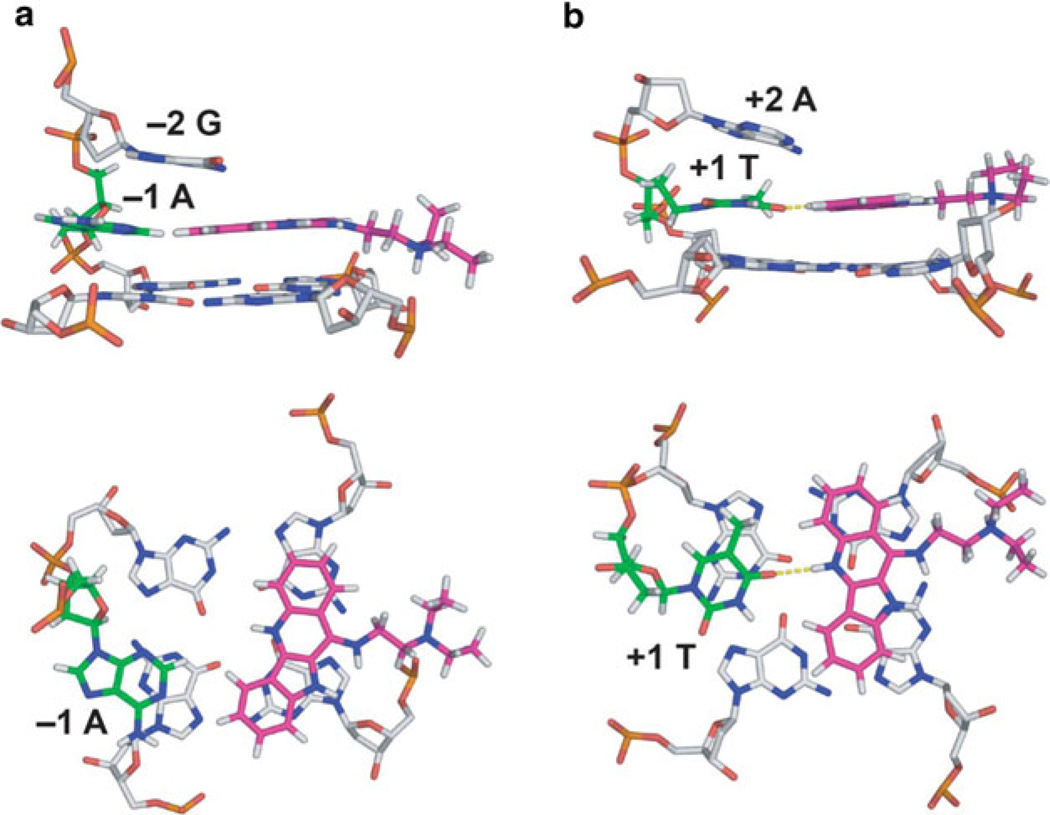
Two views showing representative structures of the 2:1 complex of quindoline-i and the c-MYC G-quadruplex are shown in Fig. 5. The quindoline-i-bound c-MYC G-quadruplex adopts the same folding pattern as that of the free DNA (see Fig. 4b) [25]. Instead of stacking on the external G-tetrads, both quindoline-i molecules bind to the c-MYC G-quadruplex in an “induced-fit” manner. Upon drug binding, both flanking sequences of the c-MYC G-quadruplex undergo an unexpected and large conformational change to assemble new capping structures. At both ends of the c-MYC G-quadruplex, the quindoline-i molecule stacks with just two of the four guanines of each external tetrad. Furthermore, the +1 thymine flanking base at the 3′-end and the −1 adenine flanking base at the 5′-end are recruited to form a quasitriad plane (Fig. 6). The +2 adenine and −2 guanine flanking residues wrap over the newly formed quasi-triad planes at each end (Fig. 6). This solution structure for the induced intercalated triad pocket shows a reorientation of the flanking sequence with the external tetrad and quindoline-i.
Fig. 5.
A representative model of the NMR-refined 2:1 quindoline-i:c-MYC G-quadruplex complex structure from two different views, prepared using GRASP (guanine = yellow, adenine = red, thymine = blue). The quindoline-i molecules are shown as a space-filling model in green. The two potassium ions are shown as gray balls. Figure is modified, with permission, from [24], © (2011) American Chemical Society
Fig. 6.
Two different views of the drug-induced binding pockets at the 5′-end (a) and the 3′-end (b). The 3′- and 5′-end flanking bases are labeled. Figure is modified, with permission, from [24] © (2011) American Chemical Society
2.2 Different Binding Interactions Between the 3′-End and 5′-End Complexes
The 3′- and 5′-ends of the quindoline-i:c-MYC G-quadruplex complex have many features in common, including base stacking over two adjacent guanines and recruitment of either the −1 or +1 base that is aligned in the same plane as quindoline-i. However, the 3′- and 5′-ends also exhibit some important differences. For example, the 5′-face is more hydrophobic and more accessible for ligand stacking, whereas the 3′-face is more hydrophilic and less accessible for ligand stacking. Furthermore, the 5′-end complex is the more stable end. Its stability is highly dependent on the capping −2 guanine (mentioned above) since deletion of or mutation to a thymine dramatically reduces the stability of the complex (Fig. 6a). The 3′-end complex is only stable under low ionic strength, emphasizing the importance of ionic rather than stacking interactions (Fig. 6b). The origin of the differences between the two ends is likely due to inherent structural features associated with the 3′- and 5′-faces as well as the flanking sequences.
2.3 Comparisons with Other Ligand:G-Quadruplex Complexes
Most of the known ligand:G-quadruplex complex structures have been derived from telomeric sequences that form bimolecular and tetramolecular species and determined by X-ray crystallography [33]. Prior to this work, TMPyP4, in complex with the unimolecular c-MYC promoter G-quadruplex, was the only known NMR-derived ligand:G-quadruplex structure for a promoter region [34]. However, the modified c-MYC promoter sequence used in that study contains a guanine-toinosine substitution. This substitution induces a guanine-strand discontinuity at a guanine position shown to be critical for G-quadruplex formation in the wild-type mycPu27 sequence [35]. In this complex, TMPyP4 stacks over the 5′-end; however, the orientation of TMPyP4 was not resolved by NMR data, and a well-defined binding pocket was not observed. There are two dimeric telomeric G-quadruplexes in which the recruitment of a base into the plane of the ligand similar to that demonstrated by our work occurs. In these structures, a thymine residue is recruited into an in-line triad plane [36, 37]; however, because of the multimeric nature of both structures, they are less relevant to the unimolecular species described here.
2.4 Insights into the Structure-Based Design of G-Quadruplex-Interactive Compounds
The 2:1 quindoline-i:c-MYC G-quadruplex complex structure provides an important case study for the selective binding of ligands to unimolecular parallel G-quadruplexes in the promoter elements of various genes such as c-MYC, VEGF, Hif-1α, and c-KIT [38]. An important implication from our structure is that, unlike the symmetrical cyclic ligand typified by TMPyP4, asymmetric compounds that contain a smaller stacking moiety, such as quindoline-i, show more selective binding to a unimolecular parallel G-quadruplex.
The specific binding of the quindoline-i is determined by both the identity of the binding end (3′ or 5′) and the two flanking bases. In addition, the electrostatic interaction between the diethylamino group in the side chain of quindoline-i and the DNA phosphate backbone could help orient and stabilize the quindoline-i scaffold. In turn, this electrostatic interaction may pinpoint the potential location of substituents that can interact with the loops of the G-quadruplex. Small changes in the shape or electronic structure of the ligand, or in the identity of the flanking bases, may affect the precise positioning of the ligand in relation to the G-quadruplex.
On the basis of our work, we propose a two-step process for small molecule recognition of unimolecular parallel G-quadruplexes by small molecules similar to quindoline-i. First, the small molecule induces a large conformation change in the G-quadruplex flanking bases, which results in the formation of an intercalated triad pocket. Second, the substituents on the small molecule interact with the loops of the G-quadruplex Identification of this small molecule-induced rearrangement of the flanking sequence suggests that other ligands may produce their own unique binding pockets in promoter G-quadruplexes and thus provide unexpected opportunities for structure-based drug design.
3 The CA46 Allele-Specific Transcriptional Assay
While we were developing an NMR-derived structure of the quindoline-i:c-MYC G-quadruplex complex, we also sought to provide more convincing evidence for the biological role of G-quadruplexes and their drug targeting with small molecules similar to quindoline-i. Specifically, we were interested in finding evidence that the inhibition observed for ligand-mediated transcription of c-MYC was directly mediated through the G-quadruplex found in the NHE III1 of the c-MYC promoter region and not through secondary off-target effects. Probably the most exacting test used prior to the recent publication in Journal of Biological Chemistry [17] was the comparison of effects of G-quadruplex-interactive compounds in two Burkitt’s lymphoma cell lines, CA46 and Ramos. Only in the case of the Ramos cell line is the G-quadruplex retained in the translocated allele; therefore compounds that work directly through the c-MYC G-quadruplex should have a more profound effect on c-MYC transcription in this cell line than in CA46. Indeed, TMPyP4, but not TMPyP2 (its positional and less G-quadruplex-interactive isomer), appears to have this preferential effect. Likewise, quindoline-i and actinomycin also showed positive results in this assay [29, 39]. However, even in the translocated allele of the Ramos cell line, the possibility remains that the inhibitory effect on c-MYC was at least partially due to a secondary effect. To provide a more exacting system to determine whether G-quadruplex-interactive compounds work directly through the c-MYC G-quadruplex, we embarked on a project in which we attempted to (1) identify more specific c-MYC G-quadruplex-interactive compounds, (2) demonstrate in a cellular system that the downstream effects of G-quadruplex-interactive compounds are mediated directly through the c-MYC G-quadruplex and not indirectly through other G-quadruplexes or other cellular targets, and (3) demonstrate that the activating transcriptional factors were displaced from the NHE III1 as a direct consequence of drug binding to the c-MYC G-quadruplex.
3.1 Identification of More Specific c-MYC G-Quadruplex-Interactive Compounds
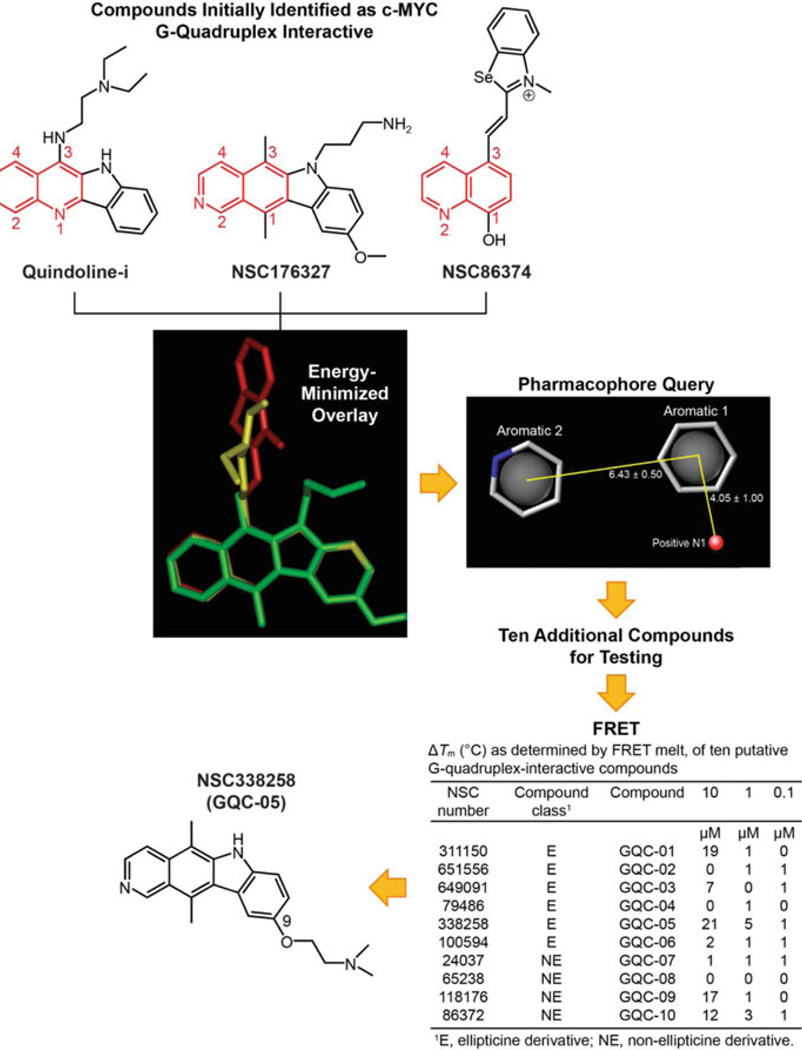
The flowchart in Fig. 7 shows the procedure used to identify GQC-05 as a compound with sufficient selectivity for the c-MYC G-quadruplex to be used in subsequent studies. Three compounds (quindoline-i, NSC176327, and NSC86374) were chosen for in silico superimposition based on their reported ability to lower c-MYC transcription or stabilize the c-MYC G-quadruplex. From the energy-minimized overlay, a pharmacophore query was generated, which was used in the NCI and ChemBridge databases to select ten additional compounds for testing in a fluorescence resonance energy transfer (FRET) melt experiment. Two drug-like property filters, MW and polar surface area, as well as synthetic accessibility were used in this selection. The FRET assay identified NSC338258 (GQC-05) as the compound that gave the maximum change in melting temperature (ΔTm).
Fig. 7.
Flow chart showing the procedure utilized to identify GQC-05 as a c-MYC G-quadruplex-interactive compound with enough selectivity to use in subsequent experiments. Initially quindoline-i, NSC176327, and NSC86374 were used in an energy-minimized overlay to give rise to the pharmacophore query. This was used as described in the text to identify NSC338258 (GQC-05) as the compound to use in the subsequent exon-specific CA46 assay and ChIP analysis
3.2 Validation of GQC-05 as a c-MYC G-Quadruplex-Binding Compound
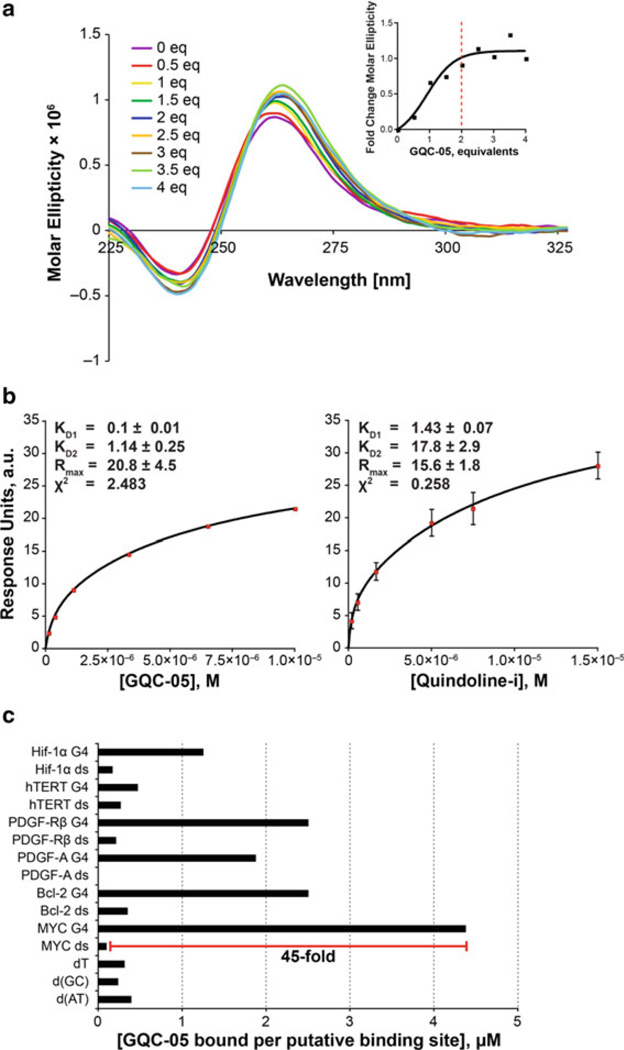
Once GQC-05 had been selected to move forward, circular dichroism (CD), competition dialysis, and surface plasmon resonance were used to determine the stoichiometry, binding parameters, and selectivity for the c-MYC G-quadruplex vs duplex and single stranded DNA as well as between different G-quadruplexes. CD demonstrated a 2:1 drug:G-quadruplex binding ratio (Fig. 8a), while the KDs for the two binding sites were 0.1 ± 0.01 and 1.14 ± 0.025 µM, which are about ten times stronger than quindoline-i. Overall, GQC-05 demonstrated more rapid binding and slower dissociation than quindoline-i (Fig. 8b). Finally, competition dialysis showed that GQC-05 had a 45-fold higher preference for the c-MYC G-quadruplex over its duplex sequence and bound with greater preference to the c-MYC G-quadruplex than to other G-quadruplexes or single-stranded and duplex DNA (Fig. 8c).
Fig. 8.
Interaction and selectivity of GQC-05. (a) CD spectra with increasing equivalents of compound demonstrate an increase in the parallel G-quadruplex structure with a peak at 262 nm. Inset: quantitation of fraction change in molecular ellipticity at 262 nm as a function of equivalents highlights a plateau at 2 eq. (b) GQC-05 demonstrated both more rapid binding and slower dissociation, with associated lower KD values for both the first and second binding sites, as compared to quindoline-i. Data represent a minimum of triplicate experiments. (c) Competition dialysis highlights that GQC-05 binds to the MYC G4 (G-quadruplex) with the highest affinity per binding site, at 45-fold selectivity over the MYC dsDNA (double-stranded DNA) sequence. Figure is modified, with permission, from [17], © (2011) American Society for Biochemistry and Molecular Biology
3.3 Demonstration in a Cellular System That the Downstream Effects of the G-Quadruplex-Interactive Compounds Are Mediated Directly Through the c-MYC G-Quadruplex and Not Indirectly Through Other G-Quadruplexes or Other Cellular Targets
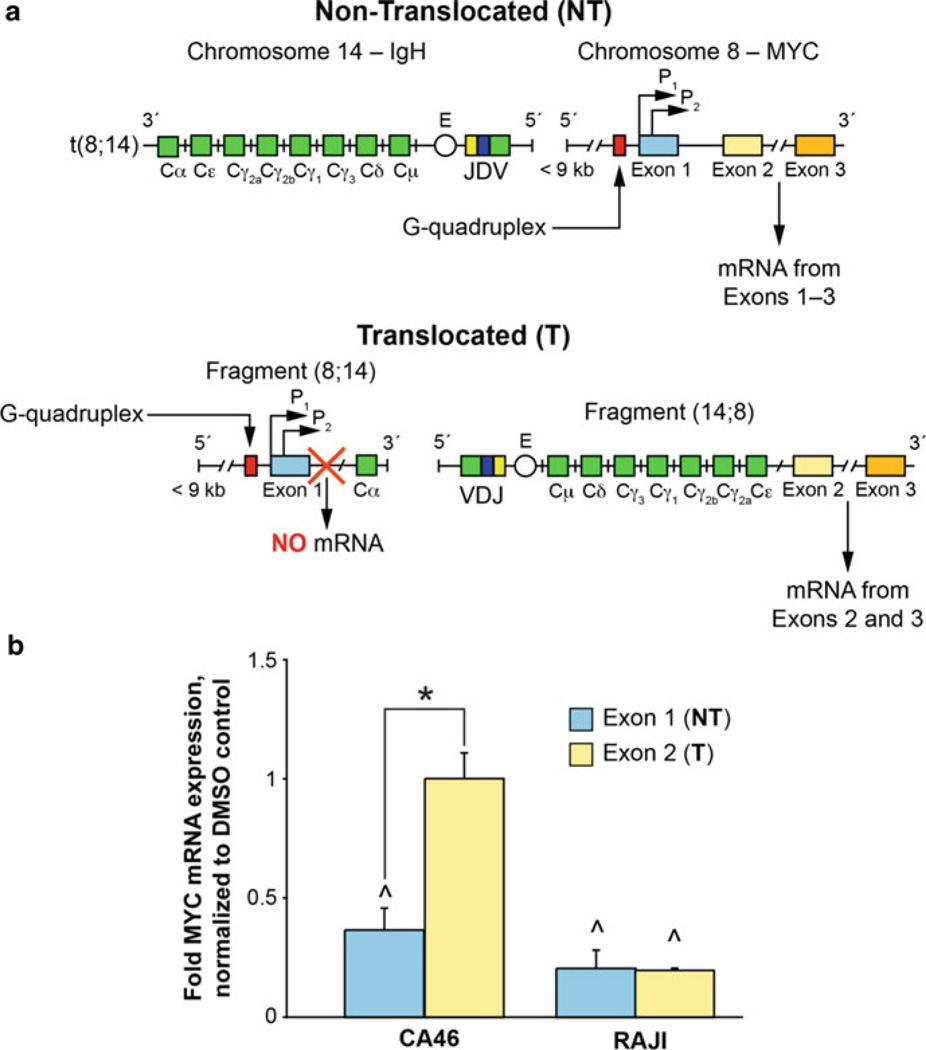
While c-MYC transcription in the translocated allele in the CA46 lymphoma cell line is not under the control of the NHE III1 and its G-quadruplex, this cell line does have the advantage that its non-translocated allele maintains the integrity of the NHE III1. Therefore, if ligands that bind to the G-quadruplex do modulate gene expression, their effect in the non-translocated allele relative to the translocated allele can be assessed. The challenge is to find a way to measure independently the c-MYC transcription mediated through the non-translocated (NT) vs translocated (T) allele. Fortunately, this can be done because exon 1 is lost from the T allele. Figure 9a shows the principle of this assay. For a ligand to be considered a specific modulator of c-MYC gene expression through the G-quadruplex, the effect should be seen exclusively through exon 1 (NT), which retains the G-quadruplex, but has little or no effect through exon 2 (T). This is exactly the result we observe for GQC-05 (Fig. 9b), where only in the NT allele do we see an effect on c-MYC transcription. Interestingly, quindoline-i did not show an exon-specific effect. These results obtained from GQC-05 are consistent with an effect mediated through the NHE III1. To further pinpoint the target to the element that contains the G-quadruplex, we performed chromatin immunoprecipitation (ChIP) assays.
Fig. 9.
The exon-specific effect of GQC-05. (a) Due to the reciprocal translocation between chromosomes 8 and 14, there are varying resultant MYC mRNAs produced. The NT products are normal, with a functional MYC under the control of a G-quadruplex, whereas the functional MYC produced from the fragment (14;8) on the T allele lacks G-quadruplex-mediated control. The G-quadruplex was removed, along with exon 1, and produces no known product from the fragment (8;14). Measurements of mRNAs containing exon 1 will mirror the NT allele; mRNAs containing exon 2 will show both the T and the NT products. (b) GQC-05 decreases MYC mRNA only from the NT allele in the CA46 cells where the G-quadruplex is maintained and has no effect from the T allele. In comparison, there is no exon-specific effect in the RAJI cells where both exons are produced under the mediated control of a G-quadruplex. All mRNA products were normalized to DMSO vehicle control; experiments at each time point are in triplicate; * p < 0.05 between exons; ^ p < 0.05 as compared to DMSO vehicle controls. Figure is modified, with permission, from [17], © (2011) American Society for Biochemistry and Molecular Biology
3.4 Demonstration That the Activating Transcription Factors Were Displaced from the NHE III1 as a Consequence of Drug Stabilization of the c-MYC G-Quadruplex
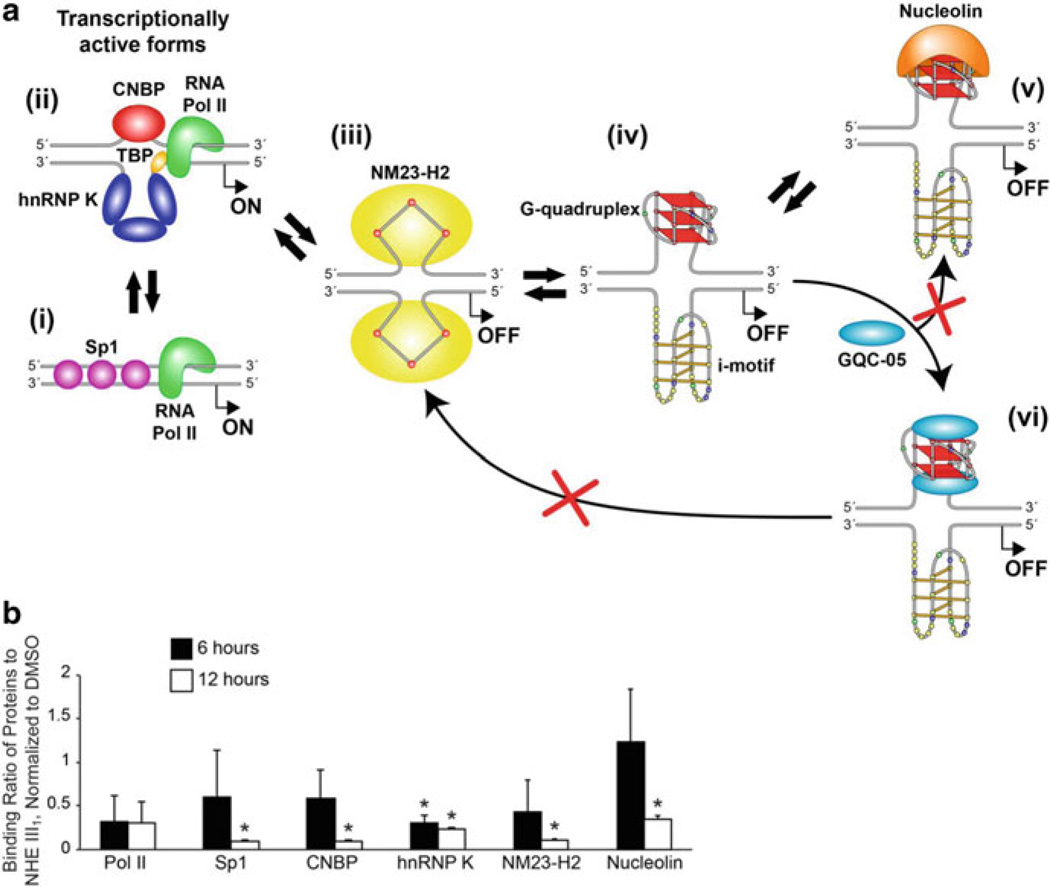
To evaluate further the specificity of the effect of GQC-05 on c-MYC transcription through the G-quadruplex in the NHE III1, we questioned whether the transcriptional factors associated with the transcriptionally active forms and the dynamic interconversion of the G-quadruplex/i-motif to duplex DNA are displaced in the NT allele in the CA46 cell line. The dynamics of the NHE III1 region within the c-MYC promoter are shown in Fig. 10a. Transcriptional factors involved in this dynamic equilibrium have been identified through a series of extensive studies [18, 40–42]. ChIP analysis with antibodies specific to the transcriptional factors hnRNP K, CNBP, and Sp1 showed that they are all significantly displaced by 12 h after incubation with GQC-05, and even at 6 h, hnRNP K is significantly reduced (Fig. 10b). In an important control experiment, no significant effects were seen on the expression of these transcriptional factors at 12 h [17]. Thus, the combined results from the CA46 exon-specific assay and the ChIP analysis provide convincing evidence that GQC-05 mediates its effects through the G-quadruplex in the NHE III1.
Fig. 10.
(a) Dynamics of the NHE III1 region within the MYC promoter. (i) Double-stranded DNA is bound by Sp1 in the transcriptionally active form, (ii) negative supercoiling opens the region to form single-stranded DNA to which CNBP and hnRNP K can bind to activate MYC. Alternatively (iv), the G-quadruplex and i-motif can form within this region, which can be unfolded by NM23-H2 (iii). In the absence of GQC-05, (v) nucleolin will cap and stabilize the G-quadruplex; however, the binding of two molecules of GQC-05 (vi) prevents this capping and stabilizes the MYC G-quadruplex, leading to transcriptional down-regulation. (b) ChIP analysis revealed a dynamic change in protein binding to the NHE III1 region within 6–12 h post-GQC-05 treatment. The binding of hnRNP K was significantly decreased within 6 h, and with the exception of RNA Pol II, which does not change, the binding of all other proteins is also decreased by 12 h. Data represent duplicate ChIP experiments; * p < 0.05 as compared to DMSO vehicle controls. Figure is modified, with permission, from [17], © (2011) American Society for Biochemistry and Molecular Biology
4 Molecular Modeling of GQC-05 and Quarfloxin Based on the Quindoline-i:c-MYC G-Quadruplex Complex
While quindoline-i did not show an exon-specific effect, the NMR solution structure of the quindoline-i:c-MYC G-quadruplex complex potentially provides a platform for understanding the binding interactions of other small molecules with the c-MYC G-quadruplex, such as GQC-05 and Quarfloxin [43]. Quarfloxin is a first-in-class G-quadruplex-interactive drug that was advanced to phase 2 clinical trials, and it has been proposed that its antitumor effect is mediated via displacement of nucleolin and subsequent binding of this protein to the c-MYC G-quadruplex [19]. The in silico docking of these molecules with the c-MYC G-quadruplex may provide insight into the factors governing intermolecular binding interactions.
4.1 Modeling Methods
The solution structure of quindoline-i with the c-MYC G-quadruplex (PDB code: 2L7V) was used as a starting model for docking studies [24]. The model for the wild-type c-MYC sequence structure was generated from this NMR structure using Insight II modeling software (Accelrys Inc., San Diego). The wild-type sequence was used throughout the modeling studies, and all charges were assigned using the consistent valence force field. Initial docking orientations were generated by a three-dimensional overlay of quindoline-i with GQC-05 and Quarfloxin. The 5′-end of the c-MYC G-quadruplex was used as the active site for small molecules. The small-molecule:c-MYC G-quadruplex complex was soaked in a 10-Å layer of TIP3P water. The entire assembly was minimized using 100,000 steps of Discover 3.0. This minimization was followed by molecular dynamics involving equilibration of 50 ps and simulations of 450 ps. Frames were collected every picosecond during the simulation phase. All the trajectories were analyzed using potential energy, and the 20 lowest potential energy frames were used to create an average structure. This average structure was then refined using 100,000 steps of minimization. This refined structure was used for calculation of interaction energy values (Table 1).
Table 1.
Interaction energy values for small molecules with the c-MYC G-quadruplex wild-type structure
| Compound | Interaction energy (kcal/mol) |
|---|---|
| Quindoline-i | −58.8 |
| GQC-05 | −89.7 |
| Quarfloxin | −112.5 |
4.2 Comparison of Binding Interactions
Docking results clearly show differences in the driving forces that determine the binding of these molecules with the c-MYC G-quadruplex. In the case of quindoline-i, the protonated nitrogen atom aligns itself with the central K+ channel while the quindoline ring interacts with the guanines of the G-tetrad via stacking. Thus, the diethylaminoethyl side chain of quindoline-i is placed in the two-base (5′-GA) double-chain-reversal loop and interacts with the phosphate backbone. As evident from the energy values, Quarfloxin exhibits a better interaction with the c-MYC G-quadruplex than quindoline-i. Quarfloxin, a fluoroquinolone analog, exhibits stacking interactions with an extended aromatic ring system intercalating between the top guanine tetrad and the 5′-end guanine, a site that is created by the movement of the 5′-end adenine. Additionally, the two side chains extend into the two single-base double-chain-reversal loops of the G-quadruplex structure (Fig. 11). Ellipticines are known to intercalate preferentially with GC-rich regions of DNA; the compound GQC-05 is a C9-substituted ellipticine analog. The pyridocarbazole ring of GQC-05 docks in an orientation similar to quindoline-i. However, the interaction of the pyridocarbazole ring with the G-tetrad is governed by stacking interactions, whereas the dimethylaminoethyloxy group in the side chain interacts with the negatively charged phosphate backbone in the thymine 19 single-base doublechain- reversal loop (Fig. 12).
Fig. 11.
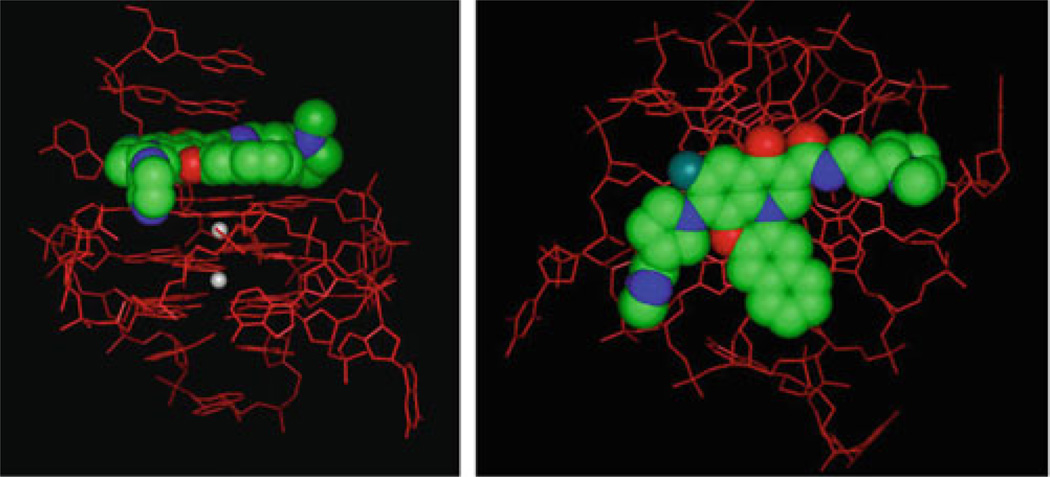
Molecular models of Quarfloxin with the c-MYC G-quadruplex structure, showing the intercalation between the top tetrad and the 5′-end flanking bases (left) and a view looking down on the top tetrad (right). Quarfloxin is shown as a space-filled model colored by atom type
Fig. 12.
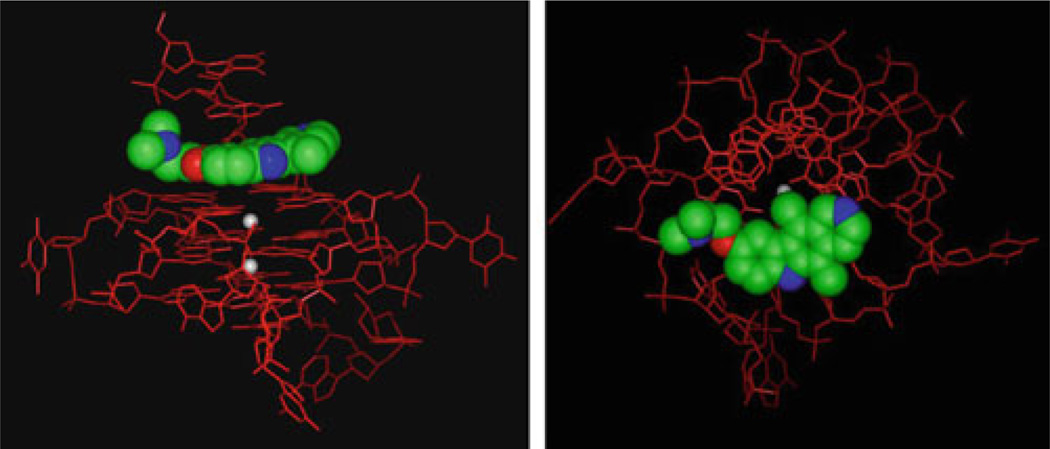
Molecular model of GQC-05 with the c-MYC G-quadruplex structure, showing the intercalation between the top tetrad and the 5′-end flanking bases (left) and a view looking down on the top tetrad (right). GQC-05 is shown as a space-filled model colored by atom type
5 Conclusions
We used NMR modeling and biological characterization to target the G-quadruplex in the c-MYC promoter in an attempt to find compounds that could down-regulate the expression of c-MYC at the transcriptional level. Our NMR work indicated a large conformational change in the flanking region of the c-MYC G-quadruplex, resulting in the formation of an induced intercalated triad pocket from the interaction between the G-quadruplex in the c-MYC promoter and the asymmetric small molecule quindoline-i. In our biological investigations, we identified GQC-05 as a potent inhibitor of c-MYC transcription. Subsequent studies using an exon-specific assay demonstrated that the effect on c-MYC transcription is mediated directly through the G-quadruplex in the NHE III1 of the c-MYC promoter. Finally, we used the NMR structure generated for the 2:1 quindoline-i:c-MYC G-quadruplex complex to glean insights about potential modes of binding for GQC-05 and Quarfloxin to the c-MYC G-quadruplex.
Acknowledgements
This research has been supported by grants from the National Institutes of Health (CA153821 [LHH] and 1S10 RR16659 and CA122952 [DY]) and the National Foundation for Cancer Research (VONHOFF0601 [LHH]). We are grateful to Dr. David Bishop for preparation, proofreading, and editing of the final version of the manuscript.
Contributor Information
Christine E. Kaiser, College of Pharmacy, University of Arizona, 1703 E. Mabel Street, Tucson, AZ 85721, USA
Vijay Gokhale, College of Pharmacy, University of Arizona, 1703 E. Mabel Street, Tucson, AZ 85721, USA; Arizona Cancer Center, 1515 N. Campbell Avenue, Tucson, AZ 85724, USA; BIO5 Institute, 1657 E. Helen Street, Tucson, AZ 85721, USA.
Danzhou Yang, Email: yangd@pharmacy.arizona.edu, College of Pharmacy, University of Arizona, 1703 E. Mabel Street, Tucson, AZ 85721, USA; Arizona Cancer Center, 1515 N. Campbell Avenue, Tucson, AZ 85724, USA; BIO5 Institute, 1657 E. Helen Street, Tucson, AZ 85721, USA.
Laurence H. Hurley, Email: hurley@pharmacy.arizona.edu, College of Pharmacy, University of Arizona, 1703 E. Mabel Street, Tucson, AZ 85721, USA; Arizona Cancer Center, 1515 N. Campbell Avenue, Tucson, AZ 85724, USA; BIO5 Institute, 1657 E. Helen Street, Tucson, AZ 85721, USA.
References
- 1.Kohn KW. Beyond DNA cross-linking: history and prospects of DNA-targeted cancer treatment—fifteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1996;56:5533–5546. [PubMed] [Google Scholar]
- 2.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 5.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 6.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 7.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 8.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 14.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J Med Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010;277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z, Gaerig V, Cui Y, Kang H, Gokhale V, Zhao Y, Hurley LH, Mao H. Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J Am Chem Soc. 2012;134:5157–5164. doi: 10.1021/ja210399h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown RV, Danford FL, Gokhale V, Hurley LH, Brooks TA. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J Biol Chem. 2011;286:41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 19.Brooks TA, Hurley LH. Targeting MYC expression through G-quadruplexes. Genes Cancer. 2010;1:641–649. doi: 10.1177/1947601910377493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michelotti GA, Michelotti EF, Pullner A, Duncan RC, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathur V, Verma A, Maiti S, Chowdhury S. Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Biochem Biophys Res Commun. 2004;320:1220–1227. doi: 10.1016/j.bbrc.2004.06.074. [DOI] [PubMed] [Google Scholar]
- 22.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai J, Carver M, Hurley LH, Yang D. Solution structure of a 2:1 quindoline–c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J Am Chem Soc. 2011;133:17673–17680. doi: 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 26.Yang D, Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guyen B, Schultes CM, Hazel P, Mann J, Neidle S. Synthesis and evaluation of analogues of 10H-indolo[3,2-b]quinoline as G-quadruplex stabilising ligands and potential inhibitors of the enzyme telomerase. Org Biomol Chem. 2004;2:981–988. doi: 10.1039/b316055f. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J-L, Lu Y-J, Ou T-M, Zhou J-M, Huang Z-S, Zhu X-F, Du C-J, Bu X-Z, Ma L, Gu L-Q, Li Y-M, Chan AS-C. Synthesis and evaluation of quindoline derivatives as G-quadruplex inducing and stabilizing ligands and potential inhibitors of telomerase. J Med Chem. 2005;48:7315–7321. doi: 10.1021/jm050041b. [DOI] [PubMed] [Google Scholar]
- 29.Ou TM, Lu YJ, Zhang C, Huang ZS, Wang XD, Tan JH, Chen Y, Ma DL, Wong KY, Tang JC, Chan AS, Gu LQ. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J Med Chem. 2007;50:1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 30.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 31.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 32.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J Am Chem Soc. 1999;121:3561–3570. [Google Scholar]
- 33.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr Opin Struct Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat Chem Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell NH, Parkinson GN, Reszka AP, Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. J Am Chem Soc. 2008;130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 37.Haider SM, Parkinson GN, Neidle S. Structure of a G-quadruplex–ligand complex. J Mol Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 38.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang HJ, Park HJ. Novel molecular mechanism for actinomycin D activity as an oncogenic promoter G-quadruplex binder. Biochemistry. 2009;48:7392–7398. doi: 10.1021/bi9006836. [DOI] [PubMed] [Google Scholar]
- 40.Dexheimer TS, Carey SS, Zuohe S, Gokhale VM, Hu X, Murata LB, Maes EM, Weichsel A, Sun D, Meuillet EJ, Montfort WR, Hurley LH. NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III1. Mol Cancer Ther. 2009;8:1363–1377. doi: 10.1158/1535-7163.MCT-08-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González V, Hurley LH. The c-MYC NHE III1: function and regulation. Annu Rev Pharmacol Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 42.González V, Hurley LH. The C-terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry. 2010;49:9706–9714. doi: 10.1021/bi100509s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]