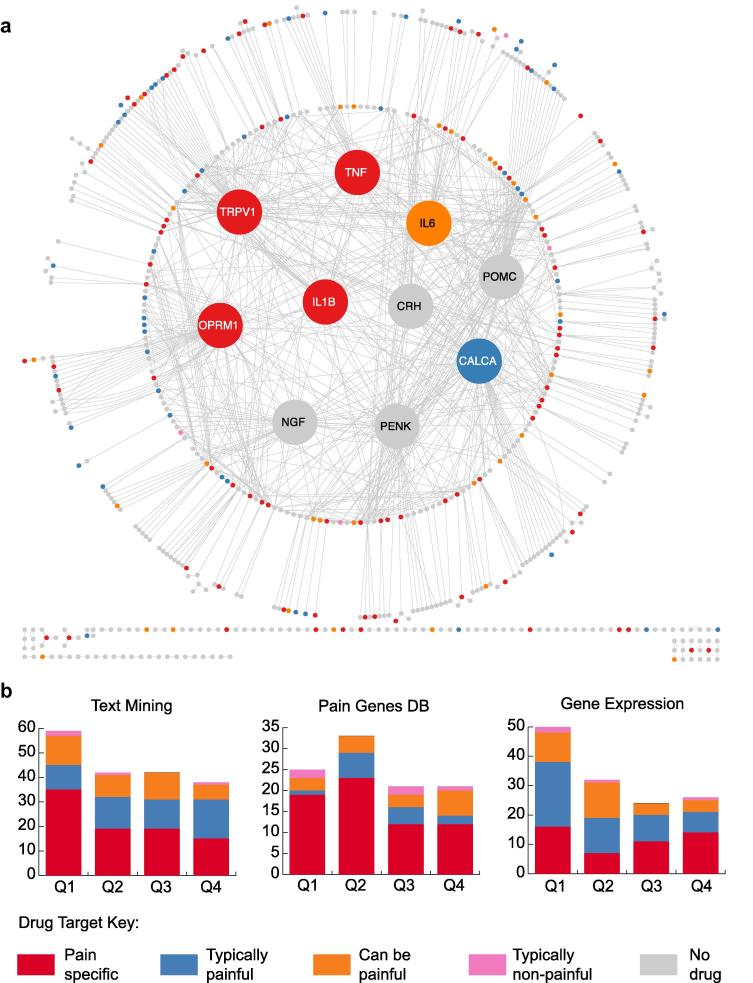
Fig. 3.
Drug targets in the pain interaction network. (a) Drug targets are color coded by the contribution of pain to their primary indication (see Methods), as indicated in the key. The 10 most enriched nodes are enlarged and moved into the center for clarity. (b) Drug target profiles of each pain network. Proteins from each dataset are ranked by their enrichment P value and binned into quartiles. Numbers of associated drugs that target proteins in each quartile are then indicated. There is a significant relationship between the enrichment of a node in the text-mined network and the likelihood of it being a drug target for a pain specific indication (χ2 test for trends in proportions, P = .002). However, neither the Pain Genes DB network nor the gene expression data show the same significant trend (P = .05 and P = 0.9, respectively).