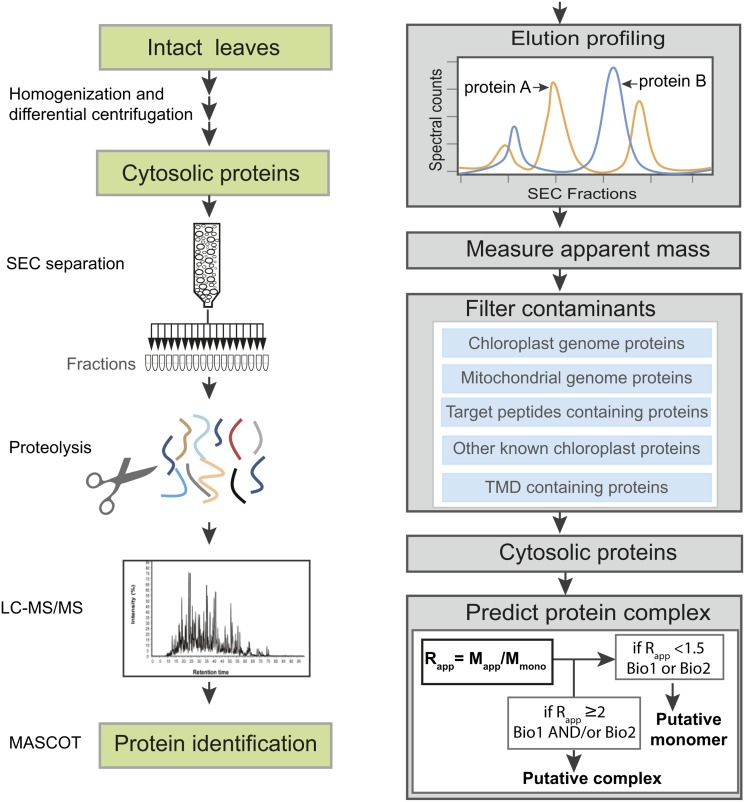
Figure 1.
Schematic Overview of the Experimental Workflow.
Proteins were extracted from intact Arabidopsis leaves under native conditions by Polytron grinding and cytosolic proteins were enriched by differential centrifugation. After separation by SEC, proteins were identified by LC-MS/MS and MASCOT database searches. Relative abundances of proteins were estimated using spectral counts, and the peak elution fraction was used to calculate apparent molecular masses, from which multimeric protein assemblies were inferred. Mapp, apparent molecular mass; Mmono, predicted molecular mass of monomer; Rapp, the ratio of the Mapp to the Mmono; Biol1, biological replicate 1; Biol2, biological replicate 2. See Methods for additional details.