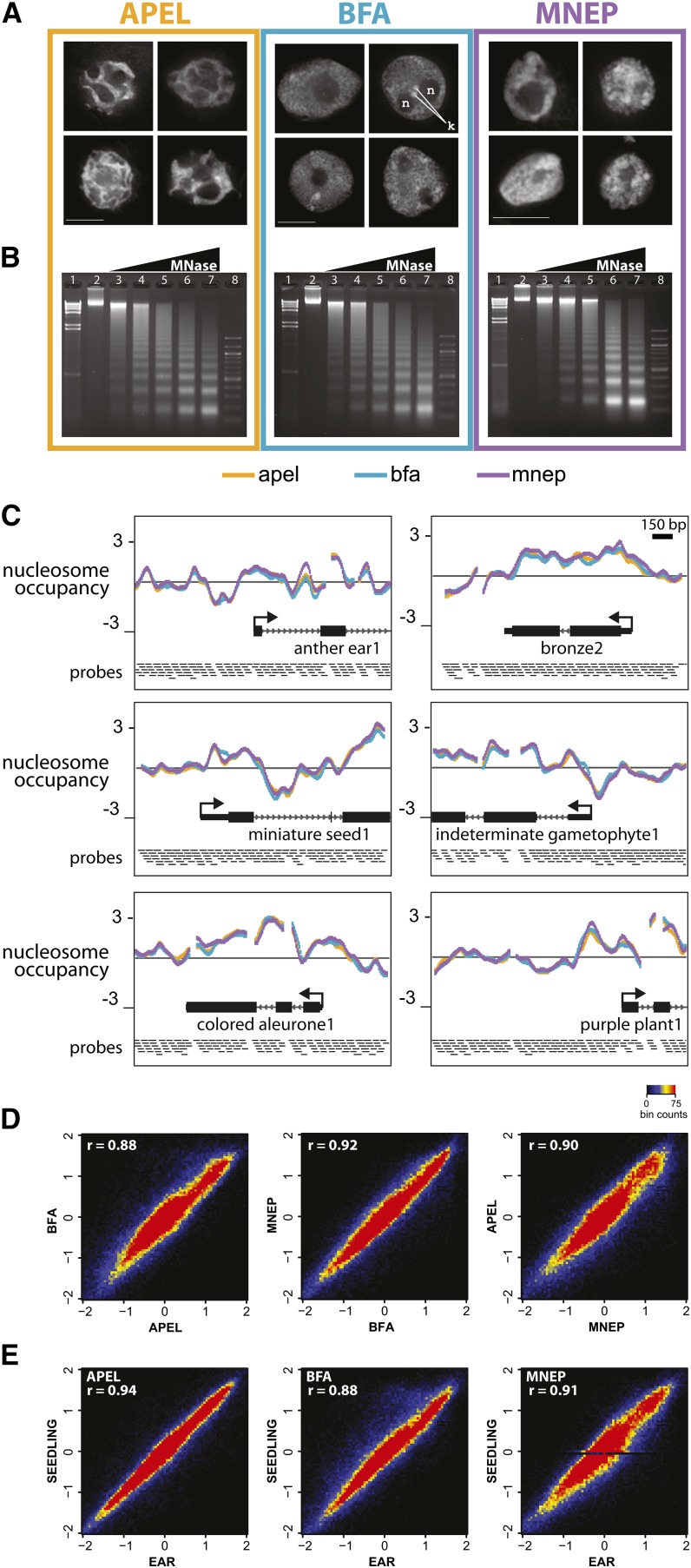
Figure 1.
Different Fixation Buffers Preserve Nuclear Morphologies to Different Degrees, but Yield Comparable Nucleosome Occupancy Profiles.
Developing ear tissue was formaldehyde-fixed and isolated in three different buffers (APEL, BFA, and MNEP), and nuclei were analyzed by three-dimensional deconvolution microscopy (A), MNase digestion products (B), and microarray-based nucleosome occupancy profiles ([C] to [E]).
(A) Single-optical middle sections of deconvolved three-dimensional data sets showing the differences in preservation of nuclear shape and chromatin appearance of immature ear nuclei among the three buffers examined. ; n, nucleolus; k, knobs. Bar = 10 μm.
(B) Developing ear nuclei were treated with titrated amounts of MNase and DNA was subjected to 1% agarose gel electrophoresis.
(C) Nucleosome occupancy plots at six example genes. Nucleosome occupancy is measured as the log2 ratio of nucleosomal/genomic fluorescence signal on a high-density microarray that includes 400 transcription start sites.
(D) and (E) Probe-by-probe scatterplots of nucleosome occupancy scores for each pairwise buffer comparison (D) and comparison of seedling against ear for each buffer (E).