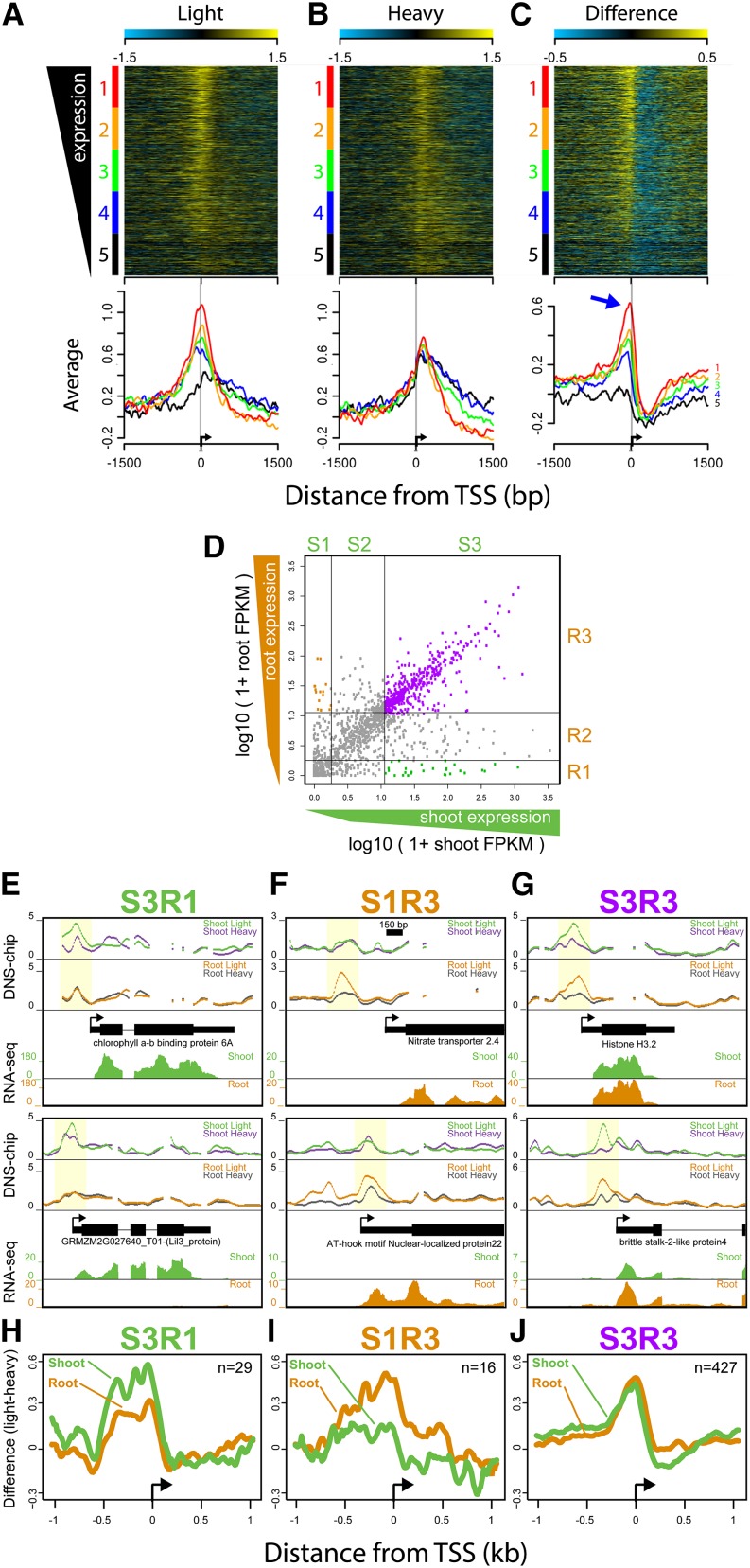
Figure 5.
MSFs Are Associated with Gene Expression Levels.
(A) to (C) Heat maps of nucleosome occupancy scores (light-digest and heavy-digest) and their difference (light-heavy) at 3 kb surrounding the 1688 TSSs included on the microarray. Gene TSSs were sorted based on their steady state gene expression levels (FPKM) and grouped into five equally sized groups (color coded at left of heat maps, from highest [red] to lowest [black]). Average scores for each of the five groups are plotted below each heat map. Blue arrow marks the −1 nucleosome region within which the difference profile covaries with gene expression levels.
(D) A scatterplot of root and shoot gene expression scores (log10(1+FPKM)) for each of the 1688 genes. Genes were grouped into three equally sized tertiles based on their gene expression level for both root and shoot, demarcated by the gray lines, which generated nine groups of genes (S1-3, R1-3).
(E) to (G) Examples DNS-chip profiles for genes highly expressed in shoots but not in roots (S3R1), genes highly expressed in roots but not shoots (S1R3), and genes highly expressed in both tissues (S3R3). Examples include nucleosome occupancy profiles for each tissue along with RNA-seq read densities for each tissue.
(H) to (J) Average difference profiles (light-heavy) at all genes within the three groups aligned by their TSS, with the number of genes in each group indicated by “n=.”