Figure 8.
Identification of CAPE Homologs in Arabidopsis and the Antipathogen Activity of AtCAPE-PR1.
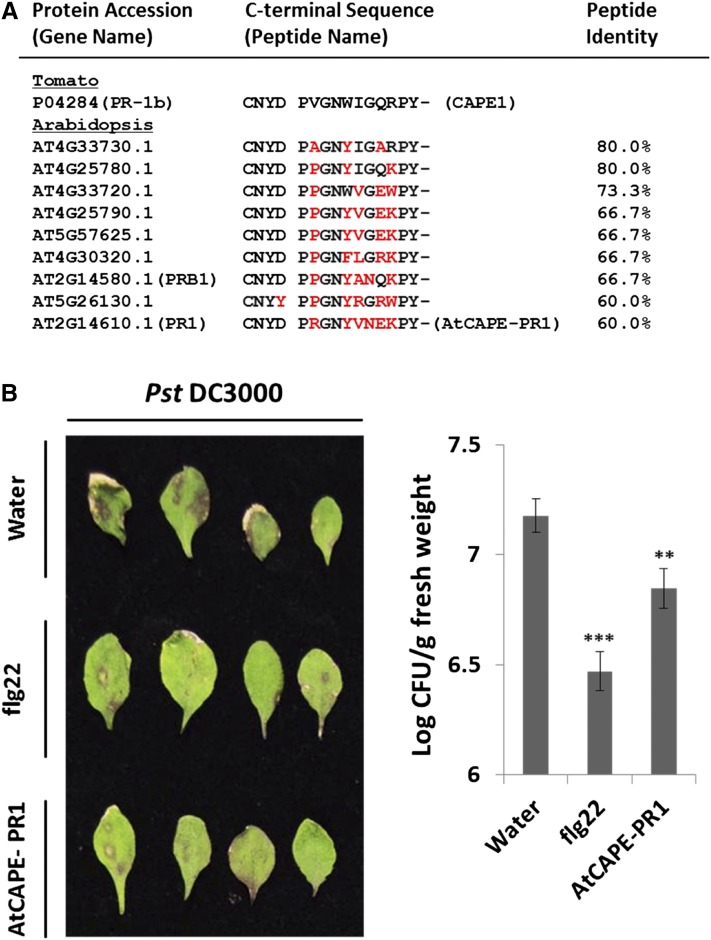
(A) The putative AtCAPE peptides derived from the CAP proteins in Arabidopsis containing a conserved cleavage (CNYx) and a signaling peptide (PxGNxxxxxPY-) motif. One of the putative AtCAPEs is derived from At-PR1, designated AtCAPE-PR1. Red characters indicated different amino acids compared with SolCAPE1.
(B) The Pst DC3000 infection phenotypes for the plants presprayed with water, 100 nM AtCAPE-PR1, or 100 nM flg22 peptide for 2 h (n = 3) prior to pathogen inoculation. The infection symptoms were observed 4 d after inoculation. The bacterial numbers were calculated 4 d after inoculation and are represented as log colony-forming units (Log CFU) per gram of leaf tissue. Data represent the means and sd of three biological samples. A statistically significant difference compared with the corresponding water-treated samples is indicated with asterisks, **P < 0.01 or ***P < 0.001, based on Student’s t test.