Fig. 1. Hyperoxia reduces intratumoral hypoxia and HIF-1α, down-regulates HIF-1α-target genes, and increases levels of HIF-1α regulating proteins including Four and a half LIM domains-1 (FHL-1).
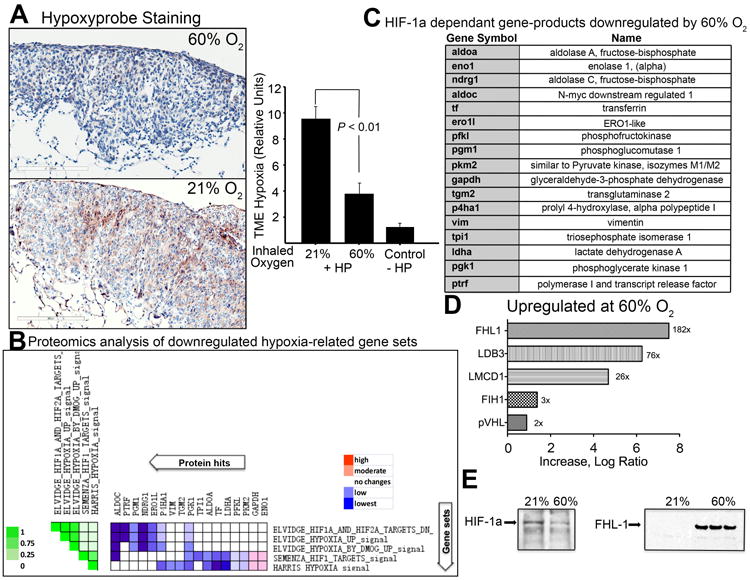
(A) Immunohistochemical staining of intratumoral hypoxia using the in vivo hypoxia marker, Hypoxyprobe. Positive Hypoxyprobe staining is shown in brown in the light micrograph. Shown is the average intensity of staining OD/mm2 (mean ± SEM, P < 0.01, n = 3). Mice bearing MCA205 pulmonary tumors breathing 60% oxygen for 3h were injected i.v. with Hypoxyprobe. (B) Expression of Hypoxia-HIF-1α dependent gene products in the TME as demonstrated by comparative whole cell proteomics analysis of known HIF-1α targets in the TME. The heat map shows the clustered genes in the leading edge subsets with the expression values represented as colors. The set-to-set graph (left) uses color intensity to show the overlap between subsets: the darker the color (on the left in green), the greater the overlap between the subsets. Leading edge analysis using GSEA software showed that five gene sets were highly correlated with the observed HIF-α regulated proteins. Only those protein sets that fall into < 0.25 false discovery rate (FDR) and were significant in the GSEA output are shown, as described previously (SI, Ref. 4-6). (C) The full names of hypoxia-regulated proteins identified by whole cell proteome analysis. (D) Proteomics analysis demonstrate HIF-1α regulating proteins (FIH, VHL, and FHL-1) are increased in tumors from mice breathing 60% oxygen. Tumor sections representing hypoxic regions (HIF-1αhigh) from mice breathing 21% O2 were compared to normoxic regions of the TME in mice breathing 60% oxygen. (E) Immunoblot assays from tumors of mice breathing 21% or 60% oxygen. Shown are representative samples of HIF-lα (left panel) and FHL-1 (right panel) from 5 independent assays.
