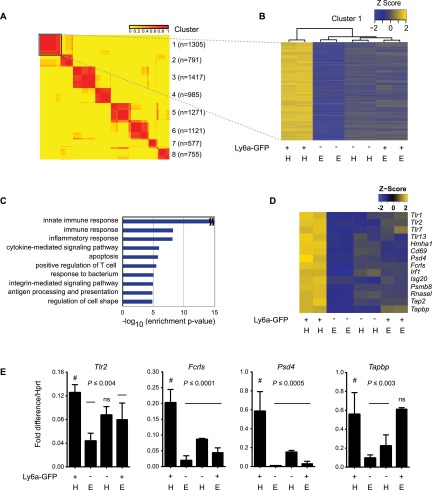
Figure 2.
Ly6a-GFP+ HCCs have an innate immune/inflammatory signature. (A) Consensus clustering of microarray data from Ly6a-GFP+ and Ly6a-GFP− HCCs and ECs. Sorts were performed as in Figure 1D. Duplicate RNA samples from each population (four total; ∼15,000 cells per sample) were analyzed by microarray. Each row represents one gene. Values in the key represent the frequency at which two genes were observed to cluster together. Cluster numbers, including gene number (n) within each set, are indicated at the right. C1 (boxed) is expanded in B. (B) Dendrogram of C1. Gene expression levels are normalized by Z-score transformation across microarray experiments. (H) HCCs; (E) ECs. (C) Top 10 enriched GO biological process terms among C1 genes. (D) Examples of innate immune/inflammatory genes with up-regulated expression in Ly6a-GFP+ HCCs. (E) qPCR analysis of several genes in D represented as fold difference relative to Hprt (n = 3, mean ± SD). Significance is by one-way ANOVA and Dunnett’s multiple comparison test with Ly6a-GFP+ HCCs as a comparator (#). (ns) Not significant.