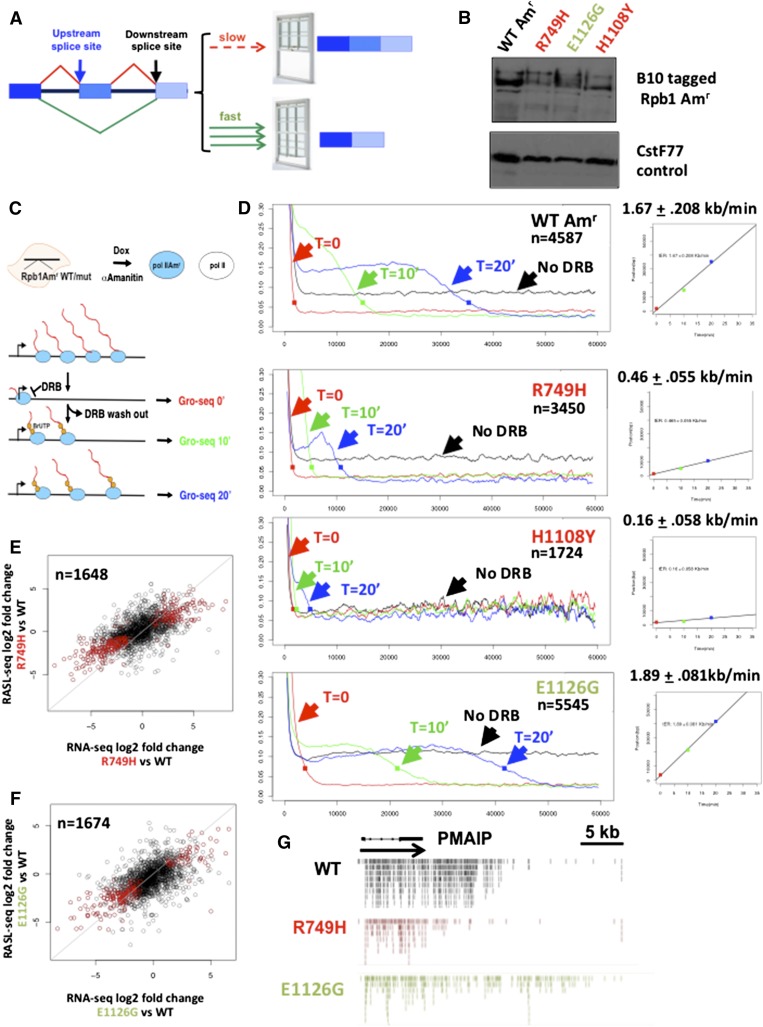
Figure 1.
Testing how elongation rate affects alternative splicing. (A) The “window of opportunity” model for how elongation rate affects alternative splicing. Slow elongation expands the window for recognition of the upstream 3′ splice site before it must compete with the downstream site, thereby favoring exon inclusion over skipping. (B) Expression of epitope-tagged human wild-type (WT) Amr and mutants with slow (red) and fast (green) elongation rates. Western blot of extracts from doxycycline-induced HEK293 Flp-in cell lines with anti B10 antibody and CstF77 loading control. (C) Scheme for measurement of elongation rates. Amanitin-resistant Rpb1 expression was induced with doxycycline, and cells were then treated with α-amanitin for 42 h to inhibit endogenous Pol II. Transcription was inhibited with DRB, which arrests Pol II at the TSS. Nuclei were harvested from DRB-inhibited cells (t = 0) and at 10 min (t = 10) and 20 min (t = 20) after washing out the inhibitor. Nuclei were processed for GRO-seq with BrUTP labeling of nascent transcripts. (D) Profiles of mean GRO-seq read counts for groups of the most highly transcribed genes with smoothing over 1-kb bins. Average elongation rates were calculated from the composite plots. (E,F) Correlation between fold change in cassette exon inclusion/exclusion in slow and fast Pol II mutants versus wild type as determined by RNA-seq and RASL-seq of α-amanitin-treated cells. Red dots correspond to exons with significant changes in inclusion (FDR < 0.05). (G) GRO-seq reads on the highly transcribed PMAIP gene that exhibits elongation-sensitive splicing (CI) (Supplemental Table 2) at the 10-min time point after DRB washout. Note that reads from the slow R749H mutant do not extend as far downstream from the TSS as wild type and that reads from the fast E1126G mutant extend further than wild type.