Figure 1.
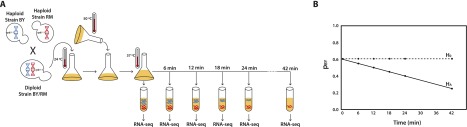
Overview of experimental design. (A) We replaced the wild-type allele of the RPO21 (also known as RPB1) gene with the temperature-sensitive rpb1-1 allele in both the BY4716 (BY) and the RM11-1a (RM) strains of S. cerevisiae (Nonet et al. 1987). We mated these two haploid strains to produce a diploid hybrid and grew the diploid to mid-log phase at the permissive temperature of 24°C. We rapidly shifted the temperature of the culture to 37°C, halting transcription. Immediately following the temperature shift, and at 6, 12, 18, 24, and 42 min after the shift, we isolated mRNA and performed RNA-seq. (B) By quantifying the relative levels of the BY and RM alleles for each gene, we estimated pBY, the proportion of transcripts derived from BY, at each time point. Under the null hypothesis (H0; dashed line) of no allele-specific differences in mRNA decay rates, pBY remains constant. Under the alternative hypothesis (HA; solid line) of allelic differences in mRNA decay, we expect pBY to change as a function of time. For the gene represented by the solid line in the example pictured, pBY decreases significantly over time, indicating that the BY allele is decaying more quickly than the RM allele of this gene.
