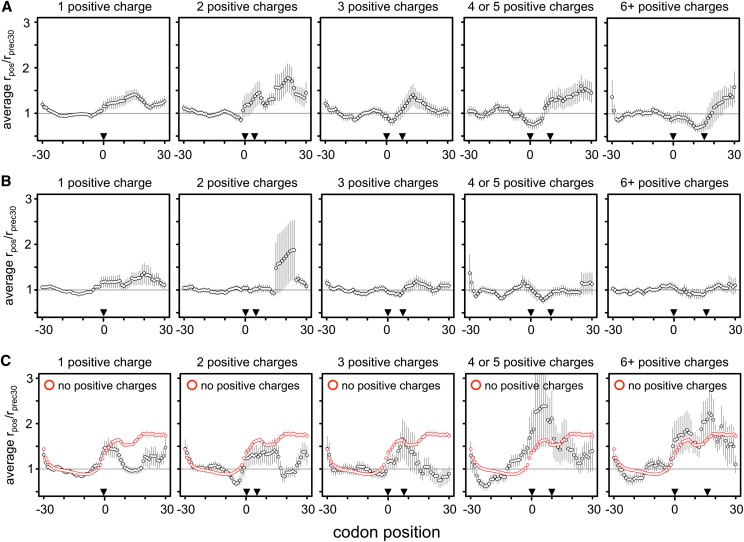
Figure 6.
No evidence of stalling at positive amino acids. We recalculated Charneski and Hurst’s (2013) Figure 5 using either the Artieri (A) or the McManus (B) data. Following the published approach, clusters of increasing numbers of positive amino acid encoding codons were identified within the range bounded by pairs of inverted triangles. The horizontal gray line indicates the average rate of translation. Error bars, ±SEM. No additive effect is observed in either high-coverage data set, in contrast to the Ingolia data (Supplemental Fig. S27); the AUCs for one, two, three, four or five, and six or more positive charge clusters were 7.89, 12.83, −0.71, −1.36, and −2.75 for the Artieri data, and 6.46, 0.08, −0.59, 0.04, and 0.09 for the McManus data, respectively. (C) The data from Charneski and Hurst’s (2013) Figure 5 (black) compared to the mean rpos/rprec30 generated from 100 random samplings of 61-codon windows devoid of any positive amino acid encoding codons (red). The average stalling pattern of windows lacking any positive charges is stronger than that observed in any of the clusters (Kruskal-Wallis rank sum test of distributions’ AUC values, P < 10−15 for all clusters except for six or more positive charges, where P = 0.02 after Bonferroni correction for multiple tests). Therefore the observed stalling effect of positive amino acids is not greater than what would be expected by chance within the Ingolia data.