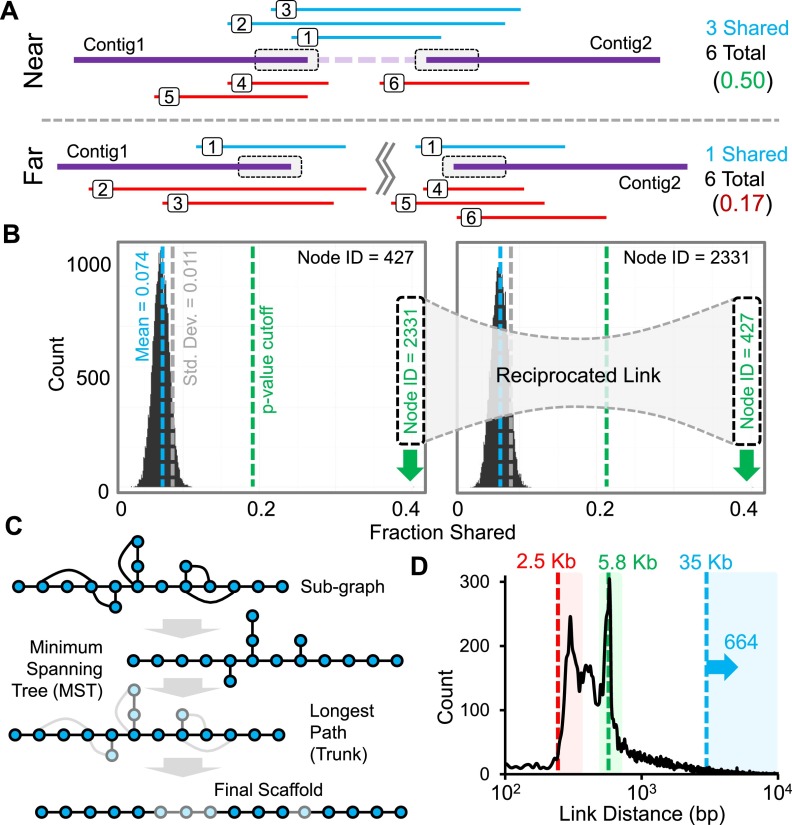
Figure 2.
fragScaff assembly method. (A) The ends of contigs in a de novo genome assembly (gray boxes) are defined as nodes, and the subsets of the 9216 CPT-seq compartments, i.e., indexed pools, containing reads that align to each node are identified. The fraction of shared compartments between every possible pair of nodes is calculated. Pairs of nodes that are truly adjacent to one another in the genome are expected to exhibit excess sharing with respect to CPT-seq compartments as a result of HMW genomic DNA fragments that bridge the gap in the de novo genome assembly. Nonadjacent pairs of nodes will co-occur in a small fraction of compartments by chance, as each contains HMW genomic fragments that cover ∼10% of the genome. (B) The fraction of shared compartments is calculated for all possible pairs of nodes, and distributions are generated for each node. Outlier nodes in each distribution are identified assuming normality and using a P-value cutoff. If a link is reciprocated, i.e., if two nodes are each outliers in the other’s distribution, it is stored as an edge. (C) Subgraphs are reduced to their minimum spanning tree (MST), and the longest path (Trunk) is found. Branches (light nodes) are then placed to produce the final output scaffold. (D) Size distribution of gaps between properly linked contigs. Boxes indicate joins spanning gaps just beyond the 2.5-kbp mate-pair library (red), ∼6 kbp L1 repeat elements (green), and joins longer than 35 kbp, which cannot be achieved via fosmid mate-pair libraries (blue; n = 664).