Figure 1.
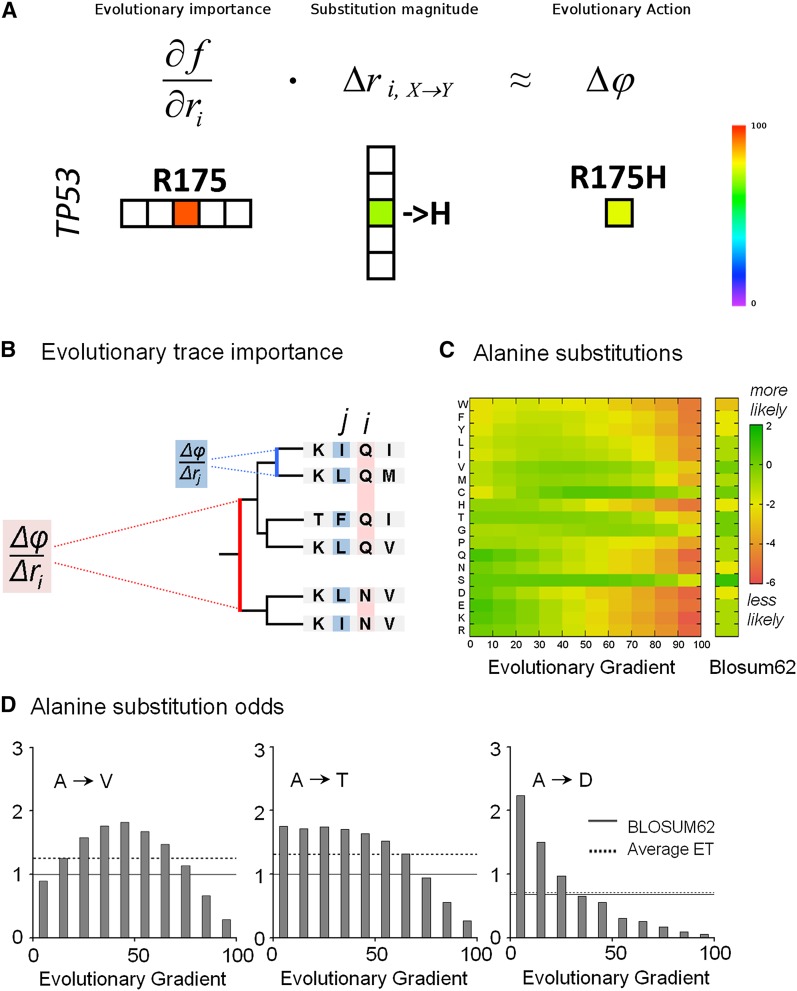
Computation of the Evolutionary Action equation. (A) An illustration of computing the Evolutionary Action of a mutation, such as the R175H in the TP53 gene, from the evolutionary importance of the residue R175 and the arginine-to-histidine substitution magnitude at that position. (B) A sequence alignment and the associated evolutionary tree show that the evolutionary fitness gradient of a protein residue, which is defined as the phenotypic fitness change due to an elementary genotypic change, will be larger (in red), or smaller (in blue), depending on the phylogenetic distance between evolutionary branches that differ at that position. Since the Evolutionary Trace ranks the functional importance of sequence positions by correlating residue variations with phylogenetic branching (Lichtarge et al. 1996; Mihalek et al. 2004), we can estimate the evolutionary fitness gradient with ET. (C) A color matrix, computed from nearly 67,000 protein sequence alignments, displays the relative substitution odds from alanine to any other amino acids (in single-letter code) depending on the evolutionary gradient decile at the mutation site (most likely substitutions are green, least likely ones are in red), and compared to the standard BLOSUM62. (D) The gradient-specific (gray bars), the nonspecific (dashed lines), and the BLOSUM62 (solid lines) substitution odds are illustrated for alanine substitutions to valine (V), threonine (T), and aspartate (D). The code is (A) alanine, (W) tryptophan, (F) phenylalanine, (Y) tyrosine, (L) leucine, (I) isoleucine, (V) valine, (M) methionine, (C) cysteine, (H) histidine, (T) threonine, (G) glycine, (P) proline, (Q) glutamine, (N) asparagine, (S) serine, (D) aspartic acid, (E) glutamic acid, (K) lysine, (R) arginine.