Abstract
Background & objectives:
Successive outbreaks of acute watery diarrhoea occurred in Talikoti and Harnal, located in Bijapur District of the southern Indian State of Karnataka, in July and August 2012, respectively. These outbreaks were investigated to identify the aetiology and epidemiology.
Methods:
Information was collected from the local population and health centres. Stool and water samples were collected from the admitted patients and their drinking water sources. Standard microbiological and PCR techniques were employed for isolation and characterization of the pathogen.
Results:
While 101 people (0.38%) were affected in Talikoti, 200 (20.94%) were affected in Harnal which is a small remote village. All age groups were affected but no death occurred. While the outbreak was smaller, longer and apparently spread by person to person contact in Talikoti, it occurred as a single source flash outbreak at Harnal. A single clone of toxigenic Vibrio cholerae O1 Ogawa biotype El Tor was isolated from the two stool samples obtained from Talikoti and subsequently from three of five stool samples obtained from Harnal indicating village to village spread of the aetiological agent. Striking similarity in antibiotic resistance profiles of these isolates with a particular strain isolated from the city of Belgaum, 250 km away, in 2010, prompted tracking the lineage of the V. cholerae isolates by DNA fingerprinting. Random amplified polymorphic DNA (RAPD) fingerprinting assay helped confirm the origin of the incriminating strain to Belgaum.
Interpretation & conclusions:
Our study reported the first twin outbreak of cholera in two remote areas of Bijapur district, Karnataka, south India. It also indicated the need for immediate preparedness to deal with such emergencies.
Keywords: Cholera, clone, epidemic, spread, Vibrio cholerae
Acute diarrhoea remains the second most prevalent communicable disease and the fourth leading cause of death in India with as many as 10762500 cases and 32218 deaths reported in 20131. Cholera, which contributes substantially to these figures, remains the most important component because of its high epidemic potential. While the disease no longer poses a threat to developed countries having appropriate standards of hygiene, it remains a challenge in India and other developing countries where access to safe drinking water and adequate sanitation facilities are often limited2. Cases of cholera have been on the rise in India and more than doubled from 1939 cases in 2006 to 5155 in 2010. While half a century ago cholera was more of a problem of the eastern parts of India3, it is increasingly being reported from southern India recently4,5,6,7,8. Outbreaks of cholera occurred in Karnataka, the second largest State of south India by area, in 2000, 2002, 2005, and 20109,10. Eighty, 117, 254, 143, and 301 cases of cholera have been reported from Karnataka in respective years during 2006-2010 which indicate that the incidence of the disease in the State has also doubled in the last five years1.
Status of cholera in Bijapur district located in the north of Karnataka is by and large unknown. The only report of cholera from Bijapur dates back to 1996 when a few cases were reported11. In this communication, we report the first outbreaks of cholera in Talikoti and Harnal, two remote locations in Bijapur district. It also traces the lineage of the incriminating strain through molecular techniques and predicts the possibility of resurgence in the near future with the intention of creating awareness among the public health authorities to help them prepare for such an emergency.
Material & Methods
Study area and demography: Talikoti is a semi-urban town with a population of 26205 located about 85 km away from the district headquarters12. It lies by the Doni, a branch of river Krishna. The town is old and has basic facilities like roads, water treatment plant and a Community Health Centre (CHC). Harnal with a population of 960 is a small village about 15 km away from Talikoti12. Most of the villagers are poor and lack treated water supply, road and sanitation facilities. Open defecation and rearing of animals are common in the area.
Case definition: A person reporting with the passage of three or more “rice-water” stools on or after July 8, 2012 was considered a suspected case of cholera. A suspected case whose stool sample on culture yielded Vibrio cholerae O1, was considered a confirmed case.
Outbreak, data acquisition and immediate control measures: On July 8, 2012, an outbreak of acute diarrhoea was detected at Talikoti when two patients were admitted to the Community Health Centre (CHC) with rice-water stools clinically suggestive of cholera. The patients were immediately put on rehydration therapy by oral or intravenous fluids depending on the extent of dehydration and were treated with tetracycline or norfloxacin and later in a few cases with iv ciprofloxacin or cefotaxime. The District Surveillance Unit Headquarters at Bijapur was alerted immediately. A few more cases were detected over the next few days with the maximum number of cases reported in a day being 10 on the 3rd and the 11th day of the report of first case. Rapid Response Team (RRT) constituted by District Surveillance Officer investigated the outbreak on July 13, 2012. Due to drying up of the Krishna river feed, the Water Treatment Plant that used to supply treated water to Talikoti was rendered defunct by early July 2012 and several borewells including a new one near the Water Treatment Plant were used as alternative drinking water sources. Water samples from these sources were collected along with stool samples from two hospitalized patients suffering from acute diarrhoea. Control measures including chlorination of water sources, distribution of halogen tablets and packets of oral rehydration salt (ORS) were carried out along with institution of information, education and communication (IEC) activities.
On August 20, 2012, an alarm was raised at the District Surveillance Unit of Bijapur with the report of as many as 85 patients suffering from acute watery diarrhoea in Harnal village. A medical team was rushed to the village immediately by the District Administration for providing health care, investigation of the outbreak and institution of public health measures. A house-to-house survey was made, villagers were interviewed and water sample was collected from a well, the only drinking water source in the village. The area around the well was found to be dirty with animal faeces scattered all around. A series of toilets with open sewage draining within 5 m from this well was noted. It was learnt that a male child who visited Talikoti for treatment of anaemia, returned to Harnal two days before the outbreak began with symptoms of diarrhoea, but did not seek medical care and recovered soon after. His case was, therefore, not recorded. A temporary clinic was opened in the village primary school for providing primary health care and treatment. Medical and paramedical staff equipped with ORS, iv fluids, and antibiotics were posted round the clock and service of an ambulance for transporting critical patients to PHC/CHC was made available. Stool samples were collected from admitted patients for laboratory investigations. ORS packets and halogen tablets were distributed and the well was disinfected with chlorine. Education on personal and public health and hygiene was also instituted for the next few days.
Clinical samples: Stool samples were randomly collected by the team of visiting health officials from admitted patients, two from Talikoti on July 13, 2012 and from five from Harnal village on August 21, 2012 for microbiological studies at BIMS and subsequently at RMRC, Belgaum. In addition, eight different strains of V. cholerae isolated by Regional Medical Research Centre (RMRC) and Belgaum Institute of Medical Sciences (BIMS) from Belgaum and surrounding areas during 2010 to 2012 were also retrieved from the strain bank of RMRC, Belgaum, and included in the study for comparison of genetic profiles10.
Isolation and identification of aetiological agent: Stool samples were collected in sterile containers and transported to the Microbiology Laboratory, BIMS and RMRC, Belgaum, within two hours on ice in raw form as well as in peptone water (PW) and alkaline peptone water (APW). After routine microscopy for detection of parasites and/or pus cells, all stool samples were enriched in PW and APW for 8 h followed by culture on blood agar, Mac Conkey agar, thiosuphate citrate bile salt sucrose (TCBS) agar, Hektoin enteric agar (HEA) at 37°C for 18 to 24 h and processed for isolation of bacterial enteric pathogens including Shigella, Salmonella, Vibrio, and diarrhoeagenic Escherichia coli following WHO guidelines13. Cultures preliminarily screened by Hanging drop technique and colonies with the characteristic appearance of V. cholerae were confirmed by biochemical tests followed by serological tests with polyvalent O1, O139 and mono-specific Ogawa and Inaba antisera (Denka Seiken, Japan). Presence of diarrhoeagenic E. coli was ruled out employing PCR14. ELISA was also performed to detect rotaviral infection15.
Antibiotic sensitivity tests: Antibiotic sensitivity tests were carried out by Kirby Bauer disk diffusion method16 using Muller-Hinton agar plates following Clinical and Laboratory Standards Institutes (CLSI), 2007 guidelines17. Antibiotic disks (Hi-Media, Mumbai) used were ampicillin (AMP 10 µg), gentamicin (GEN 10 µg), nalidixic acid (NAL 30 µg), norfloxacin (NOR 10 µg), ofloxacin (OFX 5 µg), co-trimoxazole (CoT 25 µg), tetracycline (TET 30 µg), cephalothin (CEF 30 µg), ceftriaxone (CRO 30 µg) and chloramphenicol (CHL 30 µg).
PCR assays: Multiplex PCR was performed with ctxA-tcpA primers developed by exploiting biotype specific variation of nucleotide sequences of tcpA, responsible for the expression of major subunit protein (TcpA) of toxin co-regulated pilus (TCP) of V. cholerae18. Amplification of ctxA and tcpA El Tor produces bands of 301 and 472 bp, respectively. PCR was also carried out for amplification of the rfb genes to determine their identity as O1 or O13919.
Random amplified polymorphic DNA (RAPD) fingerprinting assay: Purified genomic DNA was isolated from overnight grown cultures of V. cholerae in Luria broth (BD, USA) following cetyltrimethylammoniumbromide method20. Primer PB1 (5’-GCG CTG GCT CAG-3’) was employed in RAPD fingerprinting assay with all isolates10,21. Similarly, RAPD was also carried out with primer M16 (5’-AAA GAA GGA CTC AGC GAC TGC G-3’) to reconfirm the clonality of the isolates as described previously10,21. PCR products were electrophoresed onto 1 per cent agarose gel, with gel red dye, viewed under ultraviolet light and documented in a gel documentation system (Syngene, UK).
Collection and testing for water samples: Six water samples were collected as part of routine surveillance activity from different drinking water sources in Talikoti on July 7, 2012 and one from Harnal on August 20, 2012, and were transported to laboratory maintaining cold chain for determination of faecal contamination levels by a rapid H2S strip test (Hi-Media Laboratories, Mumbai) and by using the multiple-tube Most Probable Number (MPN) method as carried out previously7,22,. Repeat samples were collected from Talikoti on July 11, 21 and on August 24, 2012 while repeat sample from the well at Harnal was collected on August 24, 2012. One additional water sample from a household was also collected on August 24, 2012 from Harnal. Attempts were made to isolate bacterial enteric pathogens from the repeat environmental samples by enrichment in PW/APW followed by plating on selective and enrichment agar medium as mentioned earlier for stool samples. pH of water was noted during collection with a pocket pH meter.
Results
The Talikoti outbreak was detected on July 8, 2012 after two cases were reported. These two patients did not travel elsewhere prior to their infection and could not give history of close contact with any case of diarrhoea. Over the next 23 days a total of 101 cases were detected suggesting an attack rate of 0.38 per cent. The outbreak subsided on July 27, 2012 and no further case was detected. The Harnal outbreak started on August 20, 2012 and subsided within 7 days by August 26, 2012 but a total of 200 people (20.94%) were affected. No death was reported either from Talikoti or from Harnal. Vibrio cholerae was isolated from both the stool samples collected from admitted patients in Talikoti and three of five samples collected from patients in Harnal. Shigella, Salmonella or diarrhoeagenic E. coli were not isolated from any of these samples which were also negative for rotavirus by ELISA. All the five V. cholerae isolates belonged to O1 serogroup and biotype El Tor.
Antibiotic sensitivity: All the V. cholerae isolates were multiple drug resistant (MDR) showing resistance to co-trimoxazole, ampicillin, and nalidixic acid. However, these were all sensitive to tetracycline, ofloxacin, ciprofloxacin, chloramphenicol, gentamicin, ceftriaxone and cephalothin (Table).
Table.
Details of V. cholerae strains isolated during the outbreak from affected cases
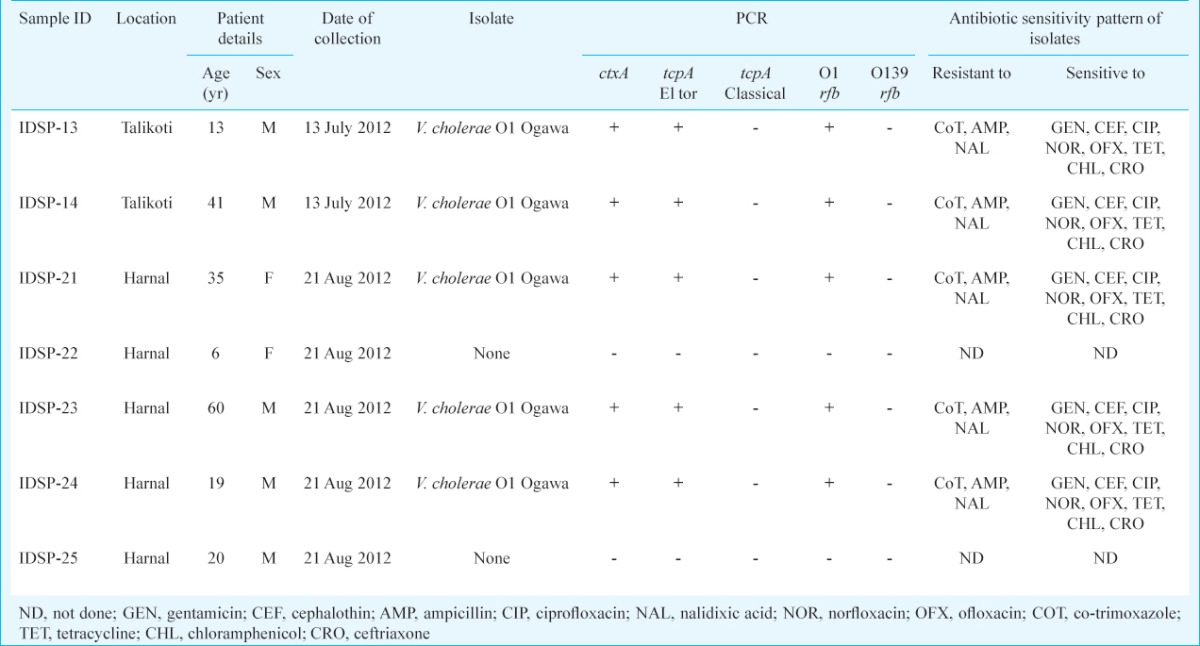
PCR assay: All isolates showed the presence of 301 bp amplicon, marker for the very potent cholera toxin gene ctxA20 and 472 bp amplicon toxin-co-regulated pilus gene tcpA for El Tor biotype19 indicating high virulence potential (Table). All isolates also showed the presence of 192 bp amplicon marker for O1rfb gene19.
RAPD fingerprinting: RAPD fingerprinting assay, revealed that the Talikoti and the Harnal outbreaks were caused by the same clone of V. cholerae O1. RAPD fingerprints of these isolates were found to be identical with one particular clone out of eight different clones of V. cholerae O1 obtained from an outbreak that occurred in Belgaum in 201010. This particular clone has also been isolated from sporadic cases in Belgaum city and nearby villages during June-August 2012 (unpublished data, RMRC, Belgaum).
Water sample analysis: Analysis of all water samples collected from Talikoti during and before the outbreak revealed gross contamination (MPN value >180/ml) with faecal coliforms. Water sample collected from tanker and Treatment Plant after resumption of service were, however, fit for consumption with no contamination indicated (MPN between 0-8/ml).
Well water at Harnal was found grossly contaminated (MPN >180/ml) with faecal coliforms but turned safe for drinking (MPN=0) after treatment.
Discussion
Our study reports two outbreaks of cholera from interior rural areas of Bijapur in the north of Karnataka, India. V. cholerae O1 Ogawa biotype El Tor was isolated as the sole aetiological agent from five of seven samples cultured. Stool samples were collected only from admitted patients who were already on antibiotics, which perhaps prevented us from isolating any organism from two cases. The identities of the isolated organisms were also confirmed by PCR based techniques. PCR also revealed the toxigenic nature of the isolates. Because of the remoteness of the villages and logistic constraints, the number of stool samples collected was less and may be a limitation for our study. Although the study did not rule out the possibility of any patient suffering from any other cause of gasteroenteritis during the outbreak, the possibility of any other aetiology seemed unlikely as the clinical symptoms of the affected persons were identical and no other pathogen was isolated from any of the stool samples. Since all age-groups were affected at both the locations, it is likely that cholera has hit these villages for the first time.
The first outbreak affected only a small fraction of people in Talikoti, lasted for 20 days and showed multiple peaks in the epidemic curve (not shown) suggesting that the outbreak was perhaps spreading from person to person rather than from any common source. The facts that only a handful of cases were reported each day and the affected people used different water sources, lended support to the above hypothesis and the possibility of any particular water-source getting contaminated with V. cholerae seemed unlikely.
The Harnal outbreak was larger compared to the Talikoti outbreak with more than 20 per cent people affected and as many as 85 people falling sick on the first day. The epidemic curve (not shown) suggested that the outbreak had a single source origin. Perhaps the faecal discharge from the anemic boy who had visited Talikoti for medical treatment two days before the Harnal outbreak might have come in contact with the well water, through the open sewers located beside the well, facilitated by rains resulting in a sudden outbreak a couple of days later. This is also supported by the fact that the well water collected on the day the outbreak began was grossly contaminated with faecal coliforms. Antibiotic sensitivity of the isolates was similar to those of the Talikoti outbreak and so were their RAPD fingerprints. Thus, the outbreak at Harnal was caused by introduction of a particular strain of V. cholerae most likely from Talikoti.
RAPD fingerprints also revealed that the outbreak strain was the same clone that was isolated two years ago from Belgaum, more than 250 km away10. While the Belgaum outbreak was caused by multiple strains of V. cholerae, this particular type was among the more sensitive and the most frequently encountered isolates. This strain has also been isolated from a few cases of cholera in Belgaum city, from Tirthkund village, 25 km south of Belgaum and from Sambra village 20 km north of Belgaum in 2012 (RMRC, Belgaum; unpublished data). No particular index case was identified for the Talikoti outbreak.
Collection of water samples before and at beginning of the outbreak was made as a part of routine surveillance by the health officials and was unfortunately not available later for attempting isolation of enteric pathogens. By the time attempts to isolate environmental isolates were made, chlorination had already been undertaken and perhaps that was the reason that no such pathogen was isolated from these samples. However, all water samples tested by routine surveillance before and at the beginning of the outbreak at Talikoti and at Harnal were found to be grossly contaminated with faecal coliforms indicating lack of proper sewage disposal and unhygienic potable water sources. Practice of open defecation and close association with domestic and farm animals have compounded the problem. Cholera epidemics are known to occur during the pre-monsoon and early monsoon periods in India when there is acute shortage of water and environmental sanitation is poor. Bijapur district has been one of the drought affected districts in Karnataka during 2012 with rain deficit of about 30 per cent reported upto mid-October23. Drying up of the Krishna water feed led to shutdown of the Talikoti Water Treatment Plant. Shortage of water coupled with poor environmental hygiene perhaps led to the outbreak.
This study reports the first outbreaks of cholera in rural areas of Bijapur District. It also highlights the usefulness of molecular epidemiological studies in tracking the origin and establishing the introduction of a particular strain of Vibrio cholerae in new locations.
Acknowledgment
Authors acknowledge the efforts of the District RRT, personnel of the District Surveillance Unit, Medical and Para-Medical staff of Talikoti and Harnal and Shri Mohan Raj, Attendant, RMRC, Belgaum, for their respective support. Support of the Government of Karnataka and Indian Council of Medical Research, New Delhi, is also acknowledged for meeting the expenses for the study.
References
- 1.National Health Profiles, Government of India. [accessed on October 14, 2012]. Available from: http://cbhidghs.nic.in/index1.asp?linkid=267.html .
- 2.World Health Organization. Cholera. [accessed on October 14, 2012]. Available from: http://www.who.int/topics/cholera/about/en/index.html .
- 3.Seal SC. The problem of cholera in India. Indian J Public Health. 1960;4:1–27. [Google Scholar]
- 4.Goel AK, Jain M, Kumar P, Sarguna P, Bai M, Ghosh N, et al. Molecular characterization reveals involvement of altered El Tor biotype Vibrio cholerae O1 strains in cholera outbreak at Hyderabad, India. J Microbiol. 2011;49:280–4. doi: 10.1007/s12275-011-0317-9. [DOI] [PubMed] [Google Scholar]
- 5.Jain M, Goel AK, Bhattacharya P, Ghatole M, Kamboj DV. Multidrug resistant Vibrio cholerae O1 El Tor carrying classical ctxB allele involved in a cholera outbreak in South Western India. Acta Trop. 2011;117:152–6. doi: 10.1016/j.actatropica.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Mandal J, Preethi V, Vasanthraja R, Srinivasan S, Parija SC. Resistance to ceftriaxone in Vibrio cholerae. Indian J Med Res. 2012;136:674–5. [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya D, Sayi DS, Thamizhmani R, Bhattacharjee H, Bharadwaj AP, Roy A, et al. Emergence of multidrug-resistant Vibrio cholerae O1 Biotype El Tor in Port Blair, India. Am J Trop Med Hyg. 2012;86:1015–7. doi: 10.4269/ajtmh.2012.11-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaji K, Okonjo PA, Thenmozhi R, Karutha Pandian S. Virulence and multidrug resistance patterns of Vibrio cholerae O1 isolates from diarrheal outbreaks of South India during 2006-2009. Microb Drug Resist. 2013;19:198–203. doi: 10.1089/mdr.2012.0127. [DOI] [PubMed] [Google Scholar]
- 9.Kanungo S, Sah BK, Lopez AL, Sung JS, Paisley AM, Sur D, et al. Cholera in India: an analysis of reports, 1997-2006. Bull World Health Organ. 2010;88:185–91. doi: 10.2471/BLT.09.073460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy S, Parande MV, Mantur BG, Bhat S, Shinde R, Parande AM, et al. Multidrug-resistant Vibrio cholerae O1 in Belgaum, South India. J Med Microbiol. 2012;61:1574–9. doi: 10.1099/jmm.0.049692-0. [DOI] [PubMed] [Google Scholar]
- 11.Peerapur BV, Srikant B, Sajjan AG, Patil SK, Mangalgi SS, Mantur BG. An outbreak of cholera in Bijapur. Indian J Med Microbiol. 1996;14:221–2. [Google Scholar]
- 12.Census data, Government of India. [Updated July 31, 2011] [accessed on October 14, 2012]. Available from: http://www.censusindia.gov.in/PopulationFinder/District_s.aspx?state_code=29 .
- 13.World Health Organization. Geneva, Switzerland: World Health Organization; Manual for laboratory investigations of acute enteric infections. WHO/CDD/83.3rev.1. 1987. [Google Scholar]
- 14.Orlandi PP, Magalhaes GF, Matos NB, Silva T, Penatti M, Nogueira PA, et al. Etiology of diarrheal infections in children of Porto Velho (Rondonia, Western Amazon region, Brazil) Braz J Med Biol Res. 2006;39:507–17. doi: 10.1590/s0100-879x2006000400011. [DOI] [PubMed] [Google Scholar]
- 15.Kang G, Arora R, Chitambar SD, Deshpande J, Gupte MD, Kulkarni M, et al. Multicenter, hospital-based surveillance of rotavirus disease and strains among Indian children aged <5 years. J Infect Dis. 2009;200(Suppl 1):S147–53. doi: 10.1086/605031. [DOI] [PubMed] [Google Scholar]
- 16.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute (CLSI) Approved standard M2-A10. Wayne, PA, USA: CLSI; 2007. Performance standards for antimicrobial disk. [Google Scholar]
- 18.Fields PI, Popovic T, Wachsmuth K, Olsvik O. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemic. J Clin Microbiol. 1992;30:2118–21. doi: 10.1128/jcm.30.8.2118-2121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keasler SP, Hall RH. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 20.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. 4th ed. New York: John Wiley; 1999. Short protocols in molecular biology. [Google Scholar]
- 21.Roy S, Dutta B, Ghosh AR, Sugunan AP, Nandy RK, Bhattacharya SK, et al. Molecular tracking of the lineage of strains of Vibrio cholerae O1 biotype El Tor associated with a cholera outbreak in Andaman and Nicobar Islands, India. Trop Med Int Health. 2005;10:604–11. doi: 10.1111/j.1365-3156.2005.01423.x. [DOI] [PubMed] [Google Scholar]
- 22.Sugunan AP, Roy S, Murhekar MV, Naik TN, Sehgal SC. Outbreak of rotaviral diarrhoea in a relief camp for tsunami victims at Car Nicobar Island, India. J Public Health (Oxf) 2007;29:449–50. doi: 10.1093/pubmed/fdm054. [DOI] [PubMed] [Google Scholar]
- 23.Karnataka State Natural Disaster Monitoring Centre, Government of Karnataka. [accessed on October 14, 2012]. Available from: http://www.dmc.kar.nic.in/Districtwise_Rainfall.pdf .


