Figure 2.
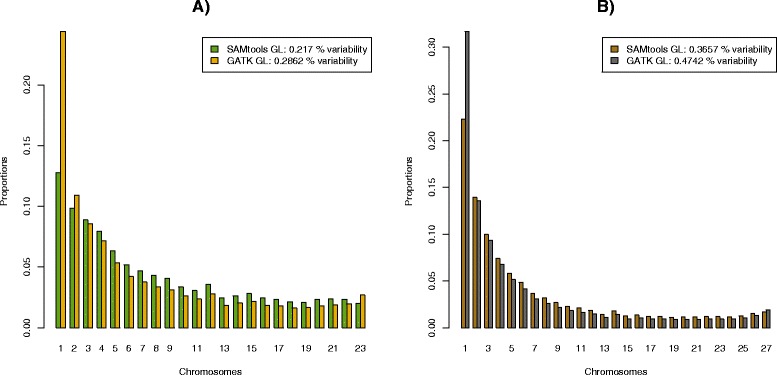
1D SFS for different GL models. SFS estimation based on a 170 megabase region from chromosome 1 using 12 CEU samples A) and 14 YRI samples B)” from the 1000 genomes project. The analysis was performed for both the GATK GL model (green, light brown) and SAMtools GL (yellow,dark brown). Notice the difference in estimated variability (proportion of variable sites) for the two GL models, with GATK GL based analyses inferring more variable sites and an associated larger proportion of low-frequency alleles. The two categories of invariable sites have been removed and the distributions have been normalized so that the frequencies of all categories sum to one for each method.
