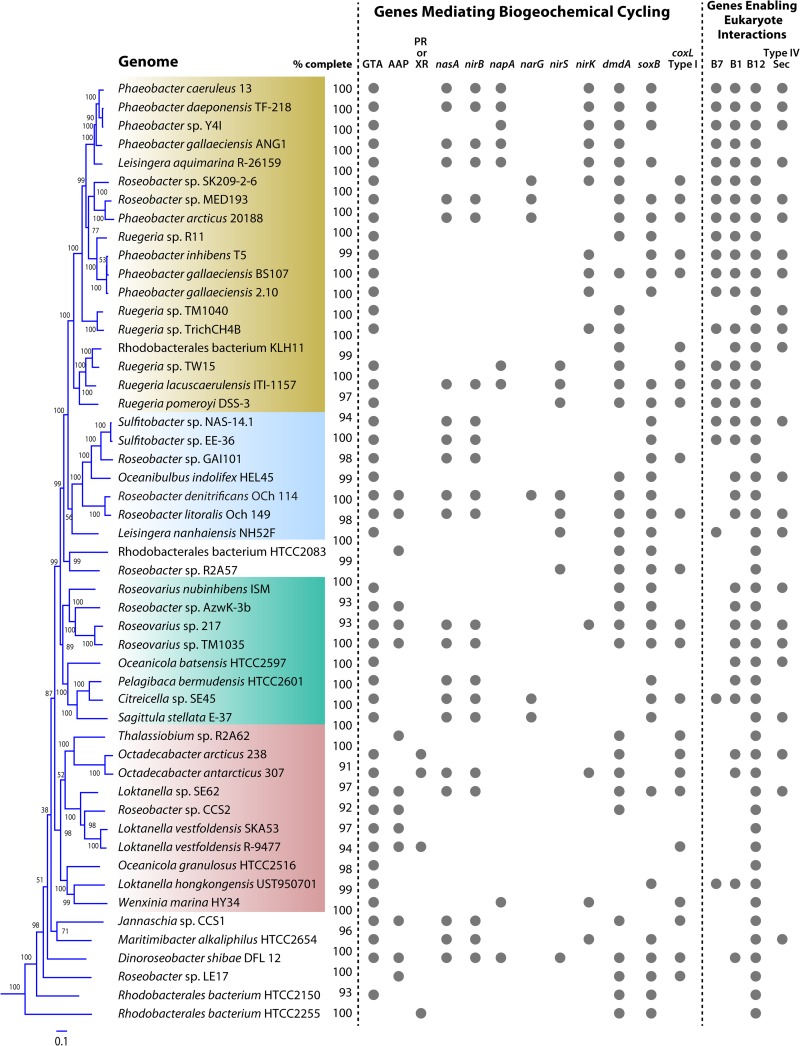
FIG 1.
Survey of select genes and metabolic pathways in 52 Roseobacter isolate genomes. % complete, estimate of genome completeness; GTA, gene transfer agent; AAP, aerobic anoxygenic photoheterotrophy; PR, proteorhodopsin; XR, xanthorhodopsin; nasA, assimilatory nitrate reductase; nirB, assimilatory nitrite reductase; napA, periplasmic dissimilatory nitrate reductase; narG, dissimilatory nitrate reductase; nirS, dissimilatory nitrite reductase; nirK, dissimilatory nitrite reductase; dmdA, dimethylsulfoniopropionate demethylase; soxB, sulfur oxidation gene; coxL type I, carbon monoxide oxidation; B7, biotin synthase; B1, thiamine synthase; B12, cobalamin synthase; Type IV Sec, type IV secretion system. Colors indicate four major clades of isolate genomes. The phylogenetic tree was constructed based on a concatenation of ∼50 single-copy conserved protein sequences using the RAxML software.