Abstract
Toxoplasma gondii and its nearest extant relative, Hammondia hammondi, are phenotypically distinct despite their remarkable similarity in gene content, synteny, and functionality. To begin to identify genetic differences that might drive distinct infection phenotypes of T. gondii and H. hammondi, in the present study we (i) determined whether two known host-interacting proteins, dense granule protein 15 (GRA15) and rhoptry protein 16 (ROP16), were functionally conserved in H. hammondi and (ii) performed the first comparative transcriptional analysis of H. hammondi and T. gondii sporulated oocysts. We found that GRA15 and ROP16 from H. hammondi (HhGRA15 and HhROP16) modulate the host NF-κB and STAT6 pathways, respectively, when expressed heterologously in T. gondii. We also found the transcriptomes of H. hammondi and T. gondii to be highly distinct. Consistent with the spontaneous conversion of H. hammondi tachyzoites into bradyzoites both in vitro and in vivo, H. hammondi high-abundance transcripts are enriched for genes that are of greater abundance in T. gondii bradyzoites. We also identified genes that are of high transcript abundance in H. hammondi but are poorly expressed in multiple T. gondii life stages, suggesting that these genes are uniquely expressed in H. hammondi. Taken together, these data confirm the functional conservation of known T. gondii virulence effectors in H. hammondi and point to transcriptional differences as a potential source of the phenotypic differences between these species.
INTRODUCTION
Toxoplasma gondii is a highly successful intracellular parasite, having infected nearly one-third of the human population (1). A member of the phylum Apicomplexa, T. gondii is primarily asymptomatic in healthy adults, but it can cause severe disease in utero and in immunocompromised humans (2–5). In rare, but nonetheless important, cases, T. gondii can cause lethal toxoplasmosis in immunocompetent adults (6). Importantly, T. gondii can infect, and cause disease in, nearly all warm-blooded animals studied to date, including multiple bird species (7–9). Such a vast host range is rare, if not unprecedented, among the members of the Apicomplexa, but the molecular basis for this feature is currently unknown and likely to be driven by multiple genetic mechanisms.
T. gondii's closest extant relative is Hammondia hammondi (10, 11), a fellow coccidian that shares the same definitive host (5). To date H. hammondi has been isolated in the field from a variety of animals, including cats, rodents, goats, and roe deer (reviewed in reference 9). Experimentally, this parasite appears to have a more restricted host range than T. gondii (e.g., H. hammondi cannot infect avian species [5]), but in contrast to the extensive work carried out on T. gondii field isolates (12, 13), there is much less information from wild H. hammondi isolates to make a definitive conclusion about the host range of this parasite.
Laboratory mice are often used as experimental models for both T. gondii and H. hammondi (9) and highlight key phenotypic differences between the species. While highly virulent T. gondii infections can be initiated in mice by multiple life stages and infection routes (14, 15), H. hammondi infections in mice can be initiated only by oocysts and infections are often asymptomatic in wild-type, immunosuppressed, and interferon gamma knockout mice (9, 16). Importantly, H. hammondi tissue cysts, which are predominantly found in muscle (whereas T. gondii tissue cysts can be found in diverse mouse tissues, including the brain), are not directly infective to mice by any route (5, 9, 16). In fact, given their morphological and antigenic similarities (17), the difference in the ability to be transmitted between intermediate hosts is used to experimentally distinguish T. gondii from H. hammondi (16).
Recently, a few key effector genes have been identified in T. gondii. These include the secreted effectors rhoptry proteins 18 and 5 (TgROP18 and TgROP5), which underlie strain-specific differences in virulence between T. gondii strains (15, 18–23). In addition, ROP16 and dense granule protein 15 (GRA15) have less dramatic, but significant, effects on virulence in the mouse and also significantly alter host cell transcription (24–27). All of the aforementioned effectors are secreted into the host cell, where they alter innate immune signaling pathways (via STAT3/6 for ROP16 and NF-κB for GRA15).
Given their high degree of genetic similarity and genomic synteny (28), T. gondii and H. hammondi provide a unique opportunity for functional comparative genomics studies. We published a genomic sequence of a strain of H. hammondi isolated from a cat in Germany (HhCatGer041) and used this sequence to determine that the H. hammondi orthologs of the major T. gondii virulence factors TgROP5 and TgROP18 are functionally conserved in H. hammondi (28). Specifically, H. hammondi ROP18 and ROP5 could complement mouse virulence defects in select T. gondii strains (28). This indicates that the mere presence of virulence factors like ROP5 or ROP18 in the H. hammondi genome is not sufficient to explain the dramatic differences in mouse virulence between these two species. In the present study, we used the HhCatGer041 genome sequence to focus on the signaling effectors GRA15 and ROP16 (25, 27). Here we show that similar to HhROP5 and HhROP18, the H. hammondi GRA15 and ROP16 orthologs are functionally conserved. By “functionally conserved,” we mean that when they are expressed in a relevant T. gondii background, they have effects on host cell signaling similar to those of their T. gondii orthologs. This provides further support for the idea that phenotypic differences between T. gondii and H. hammondi are not due to a lack of effector proteins that manipulate the host. Other mechanisms, including differences in the deployment of known effectors, the presence of currently uncharacterized “avirulence” effectors, and differences in developmental stage-switching and regulation, are more likely to underlie these phenotypic differences than gene content itself. In support of this, we also show that the transcriptomes of H. hammondi and T. gondii sporozoites are highly distinct and that their transcriptional profile differences may be a reflection of different mechanisms of growth regulation that are particularly apparent during both in vitro and in vivo growth (9).
MATERIALS AND METHODS
Parasite strains and maintenance.
Parasites were used to infect monolayers of human foreskin fibroblasts (HFFs) grown at 37°C in 5% CO2. HFFs were maintained in Dulbecco's modified Eagle medium with 10% fetal calf serum, 2 mM glutamine, and 50 μg/ml each of penicillin and streptomycin (cDMEM). RH, ME49, and CEP were used as representative type I, II, and III strains, respectively. For H. hammondi HhCatGer041 oocyst production, interferon gamma knockout mice were fed 104 H. hammondi oocysts and sacrificed 4 to 6 weeks later. For T. gondii VEG strain oocyst production, Swiss-Webster mice were used. Leg muscle (for H. hammondi) or brain (for T. gondii) tissues from infected mice were fed to young cats (10 to 20 weeks old), and feces were collected during days 7 to 11 postinfection. Unsporulated oocysts were isolated by sucrose flotation and allowed to sporulate at 4°C in 2% H2SO4 (9).
DNA isolation from H. hammondi oocysts.
Sporulated oocysts (40 million to 80 million) were washed several times in Hanks' buffered saline solution (HBSS) and then treated with 10% bleach in phosphate-buffered saline (PBS) for 30 min. After removal of all traces of bleach by multiple washes and pelleting at 800 × g in a swinging bucket rotor, pellets were resuspended in 4 ml of HBSS in a 15-ml Falcon tube, and 1 g of sterile glass beads (710 to 1,180 μM; Sigma) was added. Parasites were vortexed on high speed for 30 s, cooled on wet ice for 30 s, and then vortexed for 30 s more. After the glass beads settled, the supernatant was removed and the glass beads were washed once with HBSS; this wash was combined with the supernatant and pelleted at 800 × g. DNA was isolated from this preparation, containing sporocysts that had been freed from the oocyst wall by mechanical disruption, using the DNAzol reagent according to the manufacturer's instructions. After resuspension of the DNA pellet in sterile water, it was phenol-chloroform-isoamyl alcohol (25:24:1; Sigma) extracted and reprecipitated. For two different preparations, 8 ng of DNA was then linearly amplified using the Genomiphi DNA amplification kit and ethanol precipitated. These preparations were used for PCR amplification (described below).
Dual-luciferase reporter assays.
Putative promoters for genes derived from T. gondii and H. hammondi were compared to see how effective they were at driving firefly luciferase expression. Putative transcriptional start sites were determined using the T. gondii full-length cDNA database (http://fullmal.hgc.jp/index_tg_ajax.html [29]). For GRA15, the primers used for the H. hammondi promoter were 5′-CACCTAATAAAAATGCTCATGCACTGGT-3′ and 5′-ATCCATTGCTGAATGTTTGTTTACAAAGTG-3′ (930 bp upstream of the start codon). The primers used for the ME49 GRA15 promoter (928 bp upstream of the start codon) were 5′-CACCTAATAAAAATGATCAATACACTGGT-3′ and 5′-ATCCATTGTTGAATGCTGGTTTACAAAGTG-3′. For ROP16, the primers used for the wild-type (WT) H. hammondi promoter were 5′-CACCTTCTGGGTAGATCAGCAATAAACA-3′ (865 bp upstream of the start codon) and 5′-ATCCATCTTGCGACAAACGTAATCACAG-3′. The primers used for the wild-type ME49 ROP16 promoter (881 bp upstream of the start codon) were 5′-CACCCTCTGGGTAGAACAGCAATAGACA-3′ and 5′-ATCCATCTTGCGACAAACAAGATCACAG-3′. PCR fragments were directionally cloned into the Gateway entry vector pENTR/D-Topo (Invitrogen, Carlsbad, CA), and fragments in the proper orientation in the entry vector were recombined into a destination vector (pDestFire) containing firefly luciferase and a 3′ untranslated region (UTR) from T. gondii dihydrofolate reductase (DHFR) (30) using the Gateway LR Clonase II enzyme mix (Invitrogen) (31, 32). The forward primers included a CACC sequence required for directional cloning, while the reverse primers included an ATCCAT sequence which contains, in antisense orientation, the start codon and encodes an aspartic acid (both underlined in the primer sequences above).
We also used splicing-by-overlap extension (SOE) PCR to create chimeric ROP16 promoter luciferase constructs. To delete the 16 bp immediately preceding the putative transcriptional start site in the TgROP16 promoter, we used outer forward primer GGGGACAAGTTTGTACAAAAAAGCAGGCTCTCTGGGTAGAACAGCAATAGACA-3′ (where the underlined sequence represents a BP Clonase B1 recombination sequence) and reverse primer 5′-TGATAAATTTAATGAGTTGACTGCTCACGA-3′ to amplify the 5′ promoter sequence up to the 16 bp to be deleted. We used forward primer 5′-CATTAAATTTATCAAACATACACGATACCA-3′ and outer reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATCCATCTTGCGACAAACAAGATCACAG (where the underlined sequence represents a BP Clonase B2 recombination sequence and the start codon and additional aspartic acid codon are shown in italics) to amplify the 3′ region of the putative promoter, starting just after the 16 bp to be deleted. Similarly, we inserted the 16 bp from the TgROP16II promoter into the HhROP16 promoter. To do this, we used outer forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTCTGGGTAGATCAGCAATAAACA-3′ (B1 recombination sequence underlined as above) and reverse primer 5′-CTTCGTTTCCCATTTTAGTTGACTGCTCACGACTCG-3′ (where the 16 bp from TgROP16 are underlined) to amplify the promoter sequence up to the location of the 16-bp HhROP16 deletion. We used forward primer 5′-AAAATGGGAAACGAAG CACTAAATTTATCAAACA-3′ (where the 16 bp to be inserted is underlined as above) and outer reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCATCCATCTTGCGACAAACGTAATCACAG-3′ (where the B2 site is underlined and the start and aspartic acid codons are in italics as above) to amplify the promoter sequence immediately after the 16-bp HhROP16 deletion. Gel-purified PCR products were used in SOE PCRs with only the relevant outer forward and outer reverse primers for both TgROP16 deletion constructs and HhROP15 insertion constructs, and products were sequentially cloned into pDONR221 and pDestFire using BP and LR cloning reactions as described above. The additional nucleotide after the start and aspartic acid codons in the outer reverse primers is necessary for cloning the fragment in frame with the luciferase gene when using BP cloning for the first step in Gateway cloning. For direct comparison to the insertion and deletion promoter constructs, we also generated wild-type TgROP16 and HhROP16 promoter constructs using the outer forward and outer reverse primers, followed by an identical Gateway cloning regimen.
For luciferase assays, constructs were transfected into 15 million to 20 million parasites of the ME49 strain (between 50 and 70 μg) along with 20 μg of a vector containing Renilla luciferase driven by the TUB1 promoter, used to normalize for the efficiency of plasmid uptake by parasites in different experiments (32, 33). Parasites were harvested and lysed 24 h after transfection. Firefly and Renilla luciferase levels were measured using a dual-luciferase reporter assay system (Promega).
GRA15 cloning and characterization.
The H. hammondi GRA15 coding region plus 930 bp upstream of the start codon was amplified via PCR (forward, 5′-CACCTAATAAAAATGCTCATGCACTGGT-3′; reverse, 5′-CTACGCGTAGTCCGGGACGTCGTACGGGTATGGAGTTACCGCTGATTGTGTGCC-3′). The CACC sequence was used for directional cloning into the pENTR/D-TOPO vector (Invitrogen), and the underlined sequence in the reverse primer encodes a hemagglutinin (HA) tag and contains a stop codon. The sequence was cloned using LR Clonase recombination with the p-ATT-GRA2 destination vector (28). The p-ATT-GRA2 vector with the GRA15 insert was transfected into RHΔHXGPRT, and resistant parasites harboring the vector, which contains an HXGPRT minigene, were selected in mycophenolic acid and xanthine as described previously (34). Transgenic parasites were cloned by limiting dilution. Expression of the transgenic construct was validated using anti-HA immunofluorescence.
ROP16 cloning and characterization.
Since the H. hammondi ROP16 promoter (865 bp upstream of the predicted start codon) was not active in firefly luciferase reporter assays (see Results), the coding region was amplified using the following primers: forward, 5′-GTCCATGCATATGAAAGTGACCACGAAACGGCTT-3′, and reverse, 5′-CGTCCCATGGCTTCCGATGTGAATAAGGTTGGGTAGT-3′. For T. gondii ROP16 (from the type II strain ME49), we amplified the gene using the following primers: forward, 5′-GTCAATGCATATGAAAGTGACCACGAAAGGGCTTG, and reverse, 5′-CTGTCGTACGGGTACATCCGATGTGAAGAAAGTTCGGTAGT-3′. Underlined sites in the primer represent NsiI and NcoI sites, respectively, for HhROP16 and NsiI and BsiWI sites, respectively, for TgROP16II. Digested PCR products were ligated into the pGra-HA-HXGPRT vector (15) in frame with a C-terminal HA tag and downstream of the constitutive GRA1 promoter (35). Plasmids were transfected into ME49ΔHXGPRT, and stable transfectants were selected with mycophenolic acid and xanthine and cloned as described above.
Immunofluorescence.
Parasites were allowed to invade cell monolayers on circular 12-mm coverslips for 18 or 24 h. The cells were washed once with PBS before fixation. In most cases, the cells were fixed with 4% formaldehyde for 20 min, washed twice in PBS, and blocked in PBS supplemented with 5% bovine serum albumin (BSA) and 0.2% Triton X-100 for at least an hour. Coverslips were incubated with primary antibodies for 1 to 3 h or overnight and with fluorescent secondary antibodies for 1 h. Hoechst dye (2 μg/ml) was used for DNA visualization. Parasite strains expressing green fluorescent protein (GFP) were fixed in ice-cold methanol for 20 min. Additionally, the procedures for pSTAT6 and NF-κB staining differed from the above. For NF-κB detection, the cell monolayer was infected with parasites for 18 h, after which the cells were fixed with 3% formaldehyde for 20 min. The cells were washed twice in PBS and permeabilized with 100% ethanol for 20 min. The cells were then washed twice in PBS and blocked in PBS supplemented with 3% BSA and 5% fetal calf serum for at least an hour. The rest of the procedure followed the general protocol above. For signal quantification, 10 infected cells were chosen randomly from each coverslip and photographed with QED InVivo software (Media Cybernetics). Nuclear signal was quantified using ImageJ.
For pSTAT6 analysis, host cells seeded onto coverslips were serum starved overnight. The serum-starved monolayer was infected for 24 h, after which the cells were fixed with 4% formaldehyde for 20 min followed by ice-cold methanol for 5 min (25). The rest of the procedure followed the general protocol above. For signal quantification, 10 infected cells were chosen randomly from each coverslip and were photographed with AxioVision software (Carl Zeiss, Inc.). For each infected cell, the average nuclear pSTAT6 signal, as well as the cytoplasmic pSTAT6 signal, was quantified using ImageJ. For ROP16 nuclear localization, cells were infected for 4 h with either RHΔROP16:TgROP16I-HA or ME49:HhROP16HA at a multiplicity of infection of 10 or 8, respectively, and then fixed and stained for HA as described above. For each infection, nuclear staining was assessed qualitatively for 20 infected cells to determine the efficiency of nuclear localization. Antibodies used in this study include 3F10 (anti-HA; Roche), anti-TgSAG1 (GenWay), anti-P-Stat6 (phospho-Tyr641; Cell Signaling Technology), anti-NF-κB p65 (D14E12) XP (Cell Signaling Technology), and secondary fluorescent antibodies (Invitrogen). Monoclonal antibodies to ROP7 and GRA7 were kindly provided by Peter Bradley (UCLA) (36).
Microarray hybridizations of RNA from H. hammondi and T. gondii sporulated oocysts.
Sporulated oocysts from H. hammondi HhGER041 and T. gondii VEG were produced as described above and sporulated in 2% sulfuric acid at 4°C. Sporulated oocysts of both species were kept at 4°C for ∼20 days prior to RNA harvest. To isolate RNA, ∼80 million oocysts per species were washed with HBSS and treated with 10% bleach, and sporocysts were released using glass beads as described above for DNA isolation. Sporulation efficiency was ∼40% for both T. gondii and H. hammondi oocysts. Pellets were resuspended in TRIzol reagent (Invitrogen) by pipetting and passed serially through 25- and 27-gauge needles. Preparations were centrifuged for 1 min at 5,000 × g, supernatants were chloroform extracted, and RNA was precipitated according to the manufacturer's instructions. Pellets were resuspended in RNase-free water, extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and reprecipitated with 0.7 volume of isopropanol in the presence of 0.3 M sodium acetate (pH 5.2). Pellets were washed with 75% ethanol and resuspended in RNase-free water. Samples were labeled for array hybridization using the Illumina TotalPrep RNA amplification kit (Applied Biosystems, Carlsbad, CA). Labeled cRNA samples were hybridized using Toxoplasma 169 chips (available through the University of Pennsylvania Genomics Core Facility) (37). Three separate RNA isolations were performed for each species and hybridized separately.
Microarray data analysis.
To permit analysis of hybridization data for two species, custom Affymetrix chip description files (CDF) were generated that contained only probes that had perfect matches to sequences in the T. gondii VEG (ToxoDB) and H. hammondi HhGer041 genomes (28) using BLASTN (38). The expression module of the chip contains 11 probes for each of 8,058 T. gondii genes, and 6,294 of these genes had at least one probe that perfectly matched the T. gondii VEG and H. hammondi HhGer041 genomes. A custom script (written in Perl and available upon request) was written to generate the CDF file. Raw CEL files for the H. hammondi and T. gondii VEG oocyst hybridizations, as well as those for the complete life cycle hybridizations for T. gondii strain M4 (GEO accession number GSE32427 [39, 40]) were analyzed using the Affy package implemented in R statistical software (41), and all data were normalized (“constant” method) and log2 transformed using “median polishing” (42). To identify genes with significantly different abundances between species, we used Rank Products (43) with a false-discovery rate of 1/100 as implemented in the MeViewer Module of the TM4 microarray software suite (44). Genes were deemed to be significantly different in abundance if the adjusted P value was ≤0.01. Gene ontology (GO) analyses of differentially expressed transcripts were conducted with GeneMerge (45) as described previously (46). GO categories were deemed significantly enriched if they had adjusted P values of ≤0.05.
Using cluster analysis of transcripts of higher abundance in H. hammondi than in T. gondii VEG sporozoites along with the T. gondii M4 expression data set (39, 40), we identified two gene clusters of interest: (i) genes that were more highly expressed in H. hammondi than in T. gondii VEG oocysts that were poorly expressed in all of the life cycle stages in the T. gondii M4 data set (“Hh specific”) and (ii) genes that were more highly expressed in H. hammondi than in T. gondii VEG oocysts that were upregulated during the tachyzoite-to-bradyzoite transition in the T. gondii M4 data set (“Hh-TgCyst-high”). To identify transcripts with these profiles in an unbiased way, the entire data set was analyzed using Pavlidis template matching (PTM) (47) implemented in TM4 using a P value threshold of 1 × 10−5 and setting either the “high” values to the maximum of 1 and the “low” values to a minimum of 0. Significant genes were subjected to GO analysis as described above.
Microarray and sequence data accession numbers.
Raw data for gene expression analysis have been deposited in the Gene Expression Omnibus Database under accession number GSE61963. H. hammondi GRA15 and ROP16 gene sequences have been deposited in GenBank under accession numbers KM676011 and KM676012, respectively.
RESULTS
GRA15 from H. hammondi activates host cell NF-κB in T. gondii.
In Europe and North America several distinct, clonal genotypes of T. gondii are recognized, including types I, II, III, and IV (48–50). These strain types have significant phenotypic differences during infections in vivo and in vitro (51), and strain types I, II, and III, at least, appear to have arisen from only a few recent natural genetic crosses (52). Type II strains of T. gondii activate significantly more NF-κB than type I and III strains, and this is driven by the type II allele of TgGRA15 (27). The predicted H. hammondi GRA15 protein shares the most sequence identity with TgGRA15I and TgGRA15III (80 and 79%, respectively) and shares an 84-amino-acid insertion (relative to TgGRA15II) with TgGRA15I and TgGRA15III (see Fig. S2 in the supplemental material). However, using the neighbor-joining method based on percent identity which does not count sequence gaps (Fig. 1A), HhGRA15 is most similar to TgGRA15II, and this is reflected in the fact that HhGRA15 encodes the same amino acid as TgGRA15II at 3 of 6 sites with polymorphisms that are specific to TgGRA15II, and conversely at none of the 4 polymorphisms specific to TgGRA15III (see Fig. S2).
FIG 1.
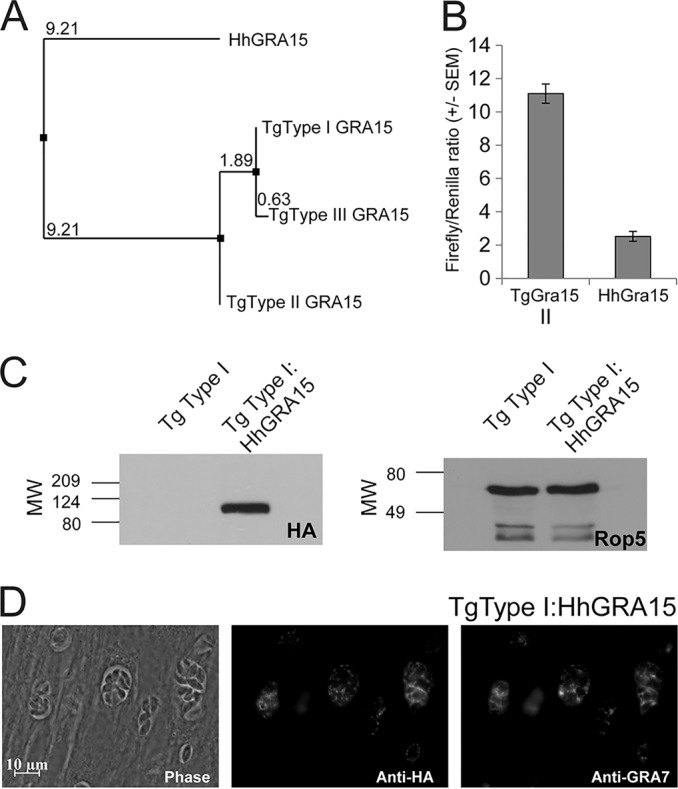
(A) Neighbor-joining tree based on percent identity between the published sequences for GRA15 from T. gondii types I, II, and III (GT1, ME49, and VEG, respectively) and that sequenced from H. hammondi HhGer041. (B) Dual-luciferase promoter assay comparing the efficacies of the putative promoters for TgGRA15II and HhGRA15. Data for two trials are shown as the ratios of firefly luciferase signal to T. gondii tubulin promoter-driven Renilla luciferase signal. (C) Western blot showing HA-tagged GRA15 (left) and loading control (ROP5). MW, molecular weight, in thousands. (D) Immunofluorescence assay showing colocalization of HA-tagged HhGRA15 with TgGRA7 in T. gondii strain RH.
We wanted to determine if HhGRA15, similar to its T. gondii type II strain ortholog, could activate the NF-κB pathway. Since to date no reliable in vitro culture system exists for H. hammondi, we used T. gondii as a surrogate for H. hammondi-derived constructs. We compared the promoter activities of HhGRA15 and TgGRA15II and found that the H. hammondi GRA15 promoter was active when expressed heterologously in T. gondii type II, though it was 4.6-fold ± 1.2-fold less active than the TgGRA15II promoter (Fig. 1B). We reasoned that the observed promoter activity for HhGRA15 was significant, in that it was ∼6,000-fold above background levels (see Table S1 in the supplemental material). To determine if HhGRA15 was functional with respect to NF-κB activation, we complemented a type I T. gondii strain (RH, which does not significantly activate NF-κB [27]) with HA-tagged HhGRA15. Importantly, HhGRA15 protein was efficiently expressed (Fig. 1C) and colocalized with the dense granule marker GRA7, indicating that it is appropriately trafficked in T. gondii (Fig. 1D). As expected, nuclear NF-κB p65 levels in T. gondii type I-infected cells were nearly equal to background, while levels in T. gondii type II-infected cells were more than 100 times greater (Fig. 2A and B). T. gondii type I expressing HhGRA15 also activated the translocation of NF-κB p65 to the host cell nucleus at a level 100 times greater than T. gondii type I (Fig. 2A and B). This activation by H. hammondi GRA15 was similar to that of T. gondii type II, suggesting that H. hammondi GRA15 acts similarly to T. gondii type II GRA15 to effect downstream induction of inflammatory and anti-apoptotic pathways via NF-κB activation (27).
FIG 2.
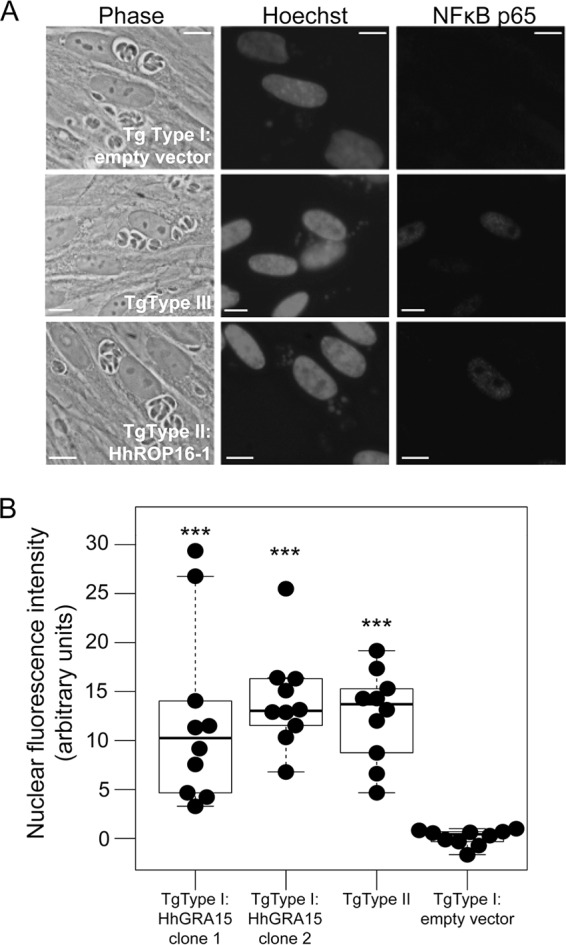
(A) Immunofluorescence analysis of NF-κB p65 activation. Infection (multiplicity of infection = 5) proceeded for 18 h before cells were fixed and stained for NF-κB p65. Like the T. gondii type II strain, T. gondii type I:HhGRA15 increases NF-κB translocation to the host cell nucleus. Scale bars: 10 μm. (B) H. hammondi HhGer041 GRA15 significantly increases the translocation of NF-κB. The mean fluorescent signal was quantified for 10 randomly chosen infected cells per strain. T. gondii type I-induced levels of NF-κB p65 were indistinguishable from the background, while levels in cells infected with T. gondii type II and T. gondii type I:HhGRA15 were similar (P > 0.05). ***, P < 0.0001 compared to T. gondii type I:empty vector.
H. hammondi ROP16 can increase STAT6 activation when expressed in T. gondii.
We identified a clear H. hammondi ortholog of T. gondii rhoptry protein 16 (ROP16) in our published H. hammondi genomic assembly and used these data to clone HhROP16. When we compared the putative promoter sequences of HhROP16 and TgROP16II, we identified a 16-bp deletion immediately preceding the predicted TgROP16I,II,III transcriptional start site (Fig. 3C; see also Fig. S3 in the supplemental material). When we fused the HhROP16 upstream region to luciferase (as for GRA15 above) and compared its activity to that of the TgROP16II promoter in a type II strain, we found that the region 865 bp upstream of the HhROP16 start codon was 25-fold less effective at driving luciferase expression than TgROP16II (Fig. 3B), and the HhROP16 upstream sequence drove luciferase expression that was only 210-fold above background levels (see Table S1). This was in contrast to that observed for the HhGRA15 promoter (above), which had activity that was ∼6,000-fold higher than the background. When we deleted the 16 bp from the TgROP16II promoter, this significantly (P = 0.002) reduced promoter activity compared to that of TgROP16WT. When we inserted 16 bp from the TgROP16 promoter into the HhROP16 promoter, promoter activity increased but was not significantly different from that of the H. hammondi WT promoter (P = 0.1). This suggests that the HhROP16 promoter is ineffective compared to the syntenic sequence in T. gondii type II, and this difference is due, at least in part, to the deletion of 16 bp relative to the T. gondii promoter 1 bp upstream of the putative transcriptional start site (Fig. 3C).
FIG 3.
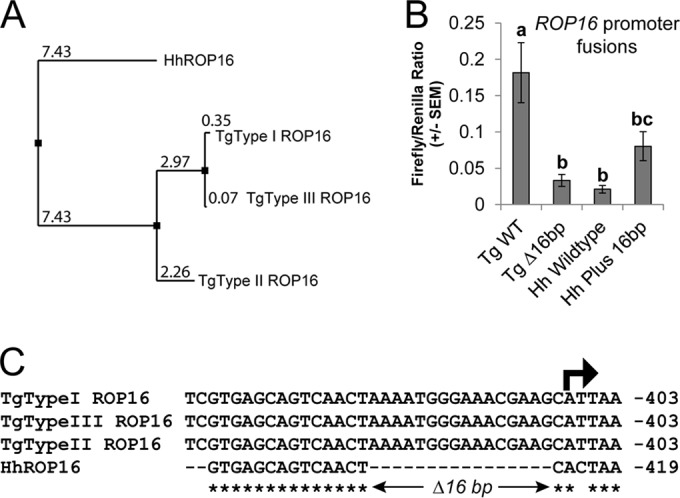
(A) Neighbor-joining tree based on percent identity between the published sequences for ROP16 from T. gondii types I, II, and III (GT1, ME49, and VEG, respectively) and that sequenced from H. hammondi HhGer041. (B) Dual-luciferase promoter assay comparing the efficacies of the putative promoters for TgROP16II WT and HhROP16 WT, as well as TgROP16II with a 16-bp deletion (Tg Δ16bp) and HhROP16 plus the 16 bp from the Toxoplasma promoters (Hh Plus 16bp). Data for 3 trials are shown as the ratios of firefly luciferase signal to T. gondii tubulin promoter-driven Renilla luciferase signal. Signal for the HhROP16 WT promoter was just slightly above the background. Means with the same letter are not significantly different (P > 0.05). (C) Alignment of the upstream region of the ROP16 gene in T. gondii types I, II, and III and H. hammondi near the predicted transcriptional start site (arrow). Asterisks indicate nucleotides that are conserved across all four sequences.
The predicted HhROP16 protein is most similar to the T. gondii type II sequence (Fig. 3A), having the same amino acid at 20 of 38 sites encoding polymorphisms that distinguish TgROP16II from TgROP16I/III (see Fig. S4 in the supplemental material). In T. gondii, ROP16 is an active kinase that directly phosphorylates both STAT3 and STAT6, resulting in their translocation to the host cell nucleus (25, 53, 54). Strain types I and III are known to more strongly activate STAT3/6 translocation than type II strains, particularly after 18 h of infection (25). The difference in this ability between these strains is driven by a polymorphism in ROP16 at residue 503 (which is a leucine in T. gondii types I and III and a serine in T. gondii type II [54]), and HhROP16 shares the leucine residue with T. gondii clonotypes I and III (see Fig. S4).
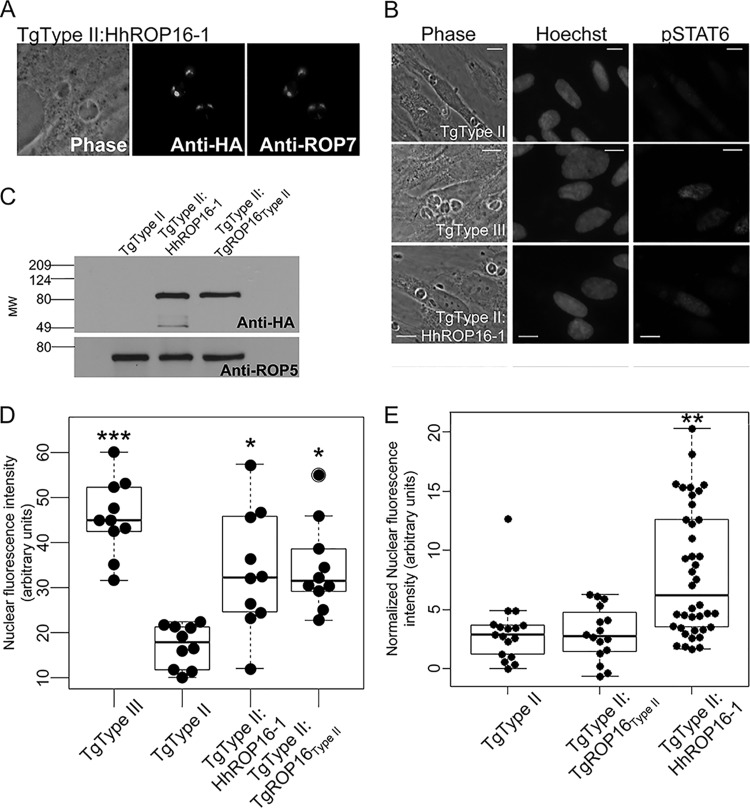
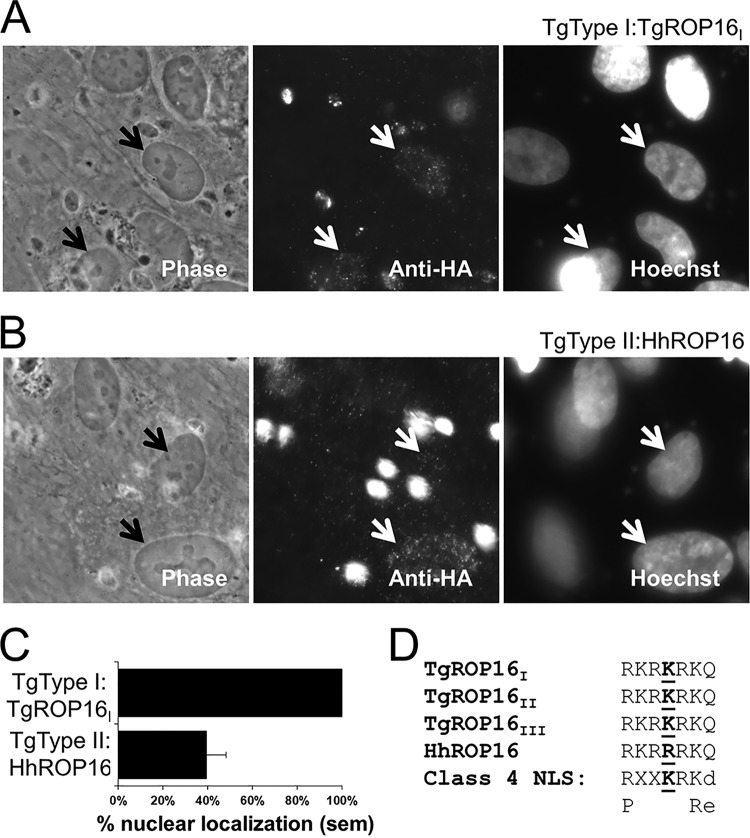
Because the H. hammondi ROP16 promoter drove weak luciferase expression, we expressed HhROP16 and TgROP16II in a type II strain of T. gondii using the constitutive TgGRA1 promoter (55). Importantly, GRA1 promoter-driven HhROP16 trafficked to the T. gondii rhoptry proteins as expected (Fig. 4A). We also examined whether, as has been shown for T. gondii ROP16 (25), H. hammondi ROP16 traffics to the host cell nucleus after infection. An important difference in the H. hammondi sequence is a single polymorphism in the putative nuclear localization signal (NLS) which meets the criteria for a monopartite class 4 NLS (Fig. 5D; see also Fig. S4 in the supplemental material) (56). To compare nuclear localizations of TgROP16 and HhROP16, we quantified the nuclear HA signal in host cells infected with either T. gondii type IΔROP16:ROP16I-HA (53) or T. gondii type II:HhROP16HA. As expected, we found that all cells infected with parasites expressing an HA-tagged version of TgROP16I had visible nuclear HA staining, while this was the case in only ∼40% of cells infected with parasites expressing HhROP16HA (Fig. 5A to C). The NLS in T. gondii ROP16 (RKRKRKQ) is required for trafficking into the host cell nucleus (25), although this trafficking is not required for activation of STAT3/6 (25). The ROP16 NLS is a class 4 NLS with the consensus sequence (R/P)XXKR(K/R)(^DE), where ^DE indicates that the terminal residue in the motif cannot be an aspartic or glutamic acid residue (25, 56). Kosugi et al. showed that when the lysine in position 4 is changed to an arginine in this motif (the putative NLS of HhROP16 has an arginine at this residue [Fig. 5D]), it reduces the efficiency of, but does not eliminate, nuclear localization of a model protein in multiple cell types (56). Our data for HhROP16 were consistent with this observation.
FIG 4.
(A) Immunofluorescence of HA-tagged HhROP16 (driven by the T. gondii GRA1 promoter) showing colocalization with the rhoptry protein ROP7. (B) Immunofluorescence analysis of pSTAT6 signaling. Infection (multiplicity of infection = 5) proceeded for 24 h before cells were fixed and stained for pSTAT6. Similar to the T. gondii type III strain, T. gondii type II:HhROP16 increases translocation of phosphorylated STAT to the host cell nucleus. (C) Western blot showing expression of HA-tagged TgROP16 and HhROP16 in T. gondii type II and the ROP5 loading control. (D) H. hammondi ROP16 significantly increases pSTAT6 signaling in T. gondii type II. Nuclear fluorescence intensity was quantified for 10 randomly selected infected cells per strain. Cells infected with type II:HhROP16 have twice the pSTAT6 signal in their nuclei as cells infected with wild-type type II. Type III induces a greater translocation of pSTAT6 to the host cell nucleus than all other strains. *, P < 0.05, and ***, P < 0.0001, compared to signal in T. gondii type II. (E) H. hammondi ROP16 significantly increases pSTAT6 nuclear localization compared to T. gondii type II and type II expressing a second copy of ROP16II. Cytoplasmic fluorescent signal was subtracted from nuclear fluorescent signal for ≥16 randomly chosen infected cells for each strain. **, P < 0.001 compared to T. gondii type II.
FIG 5.
(A) Nuclear localization of TgROP16I in cells infected with T. gondii type IΔROP16:TgROP16I 18 h postinfection. Arrows indicate two infected cells that show significant nuclear HA signal. (B) Inconsistent nuclear localization of HhROP16 in cells infected with T. gondii type II:HhROP16 18 h postinfection. Arrows indicate two infected cells, one with minimal nuclear anti-HA staining (top arrow) and another with clear nuclear anti-HA staining (bottom arrow). (C) Quantification of the percentage of infected cells exhibiting visible nuclear HA staining when infected with either T. gondii type IΔROP16:TgROP16I or T. gondii type II:HhROP16. Data shown from 20 infected cells. sem, standard error of the mean. (D) Alignment of the nuclear localization signal (NLS) in T. gondii type I, II, and III ROP16 and H. hammondi ROP16 (top 4 sequences) showing the residue at position 4 that is uniquely changed to arginine in H. hammondi ROP16. The fifth sequence is the consensus sequence for a type 4 NLS, and the last sequence shows alternative amino acids. Uppercase letters indicate permitted amino acids, and lowercase letters indicate nonpermissible amino acids.
When expressed in a T. gondii type II strain, GRA1 promoter-driven HhROP16 significantly increased STAT6 nuclear translocation compared to that of the parental type II strain (Fig. 4B and D). Additionally, we expressed the type II allele of ROP16 in type II T. gondii using the same plasmid backbone (and GRA1 promoter), and Western blotting showed similar expression levels of HA-tagged ROP16 in both T. gondii type II:HhROP16 and T. gondii type II:TgROP16II parasites (Fig. 5C). While T. gondii type II:HhROP16 significantly increased nuclear pSTAT6 compared to T. gondii type II (P < 0.001), T. gondii type II:TgROP16II infection did not significantly alter pSTAT6 localization (Fig. 4E). Therefore, expressing an additional copy of TgROP16II is not sufficient to significantly increase pSTAT6 nuclear localization, indicating that the ability of HhROP16 to increase pSTAT6 activation during infection with type II T. gondii is likely due to differences in primary amino acid sequence (including leucine 503). With respect to the lower efficiency of nuclear localization of HhROP16 than for TgROP16I, these data are consistent with what is known for TgROP16 (25): that nuclear trafficking of TgROP16 is not required for STAT6 activation and nuclear translocation. Regardless, the coding sequence of HhROP16 appears to be functionally conserved in terms of its ability to increase STAT6 activation, at least when driven by a constitutive promoter in a T. gondii type II strain, and based on its higher efficacy than for TgROP16II, it is likely most similar in function to the type I and type III ROP16 alleles.
The transcriptional profiles of H. hammondi HhCatGER041 and T. gondii VEG sporulated oocysts are distinct.
We compared transcriptional profiles of H. hammondi (HhCatGer041) and T. gondii (VEG) sporulated oocysts using T. gondii Affymetrix expression arrays (37). We used sporulated oocysts due to the difficulties in long-term in vitro culture of H. hammondi tachyzoites (9, 16). The transcriptional profiles of these species were comparable but had some striking differences (Fig. 6). Using statistical analyses, we identified 78 genes of significantly greater abundance in H. hammondi (“Hh-high” [see Fig. S5 in the supplemental material]) and 80 significantly higher in T. gondii strain VEG (“Tg-high” [see Fig. S6 in the supplemental material]). Gene ontology (GO) analysis of these gene sets did not identify any enriched GO terms in the Hh-high gene set, although 4 significant GO categories were enriched in the Tg-high set: GO:0003735 (structural constituent of ribosome; P = 2.60E−08), GO:0006412 (translation; P = 2.91E−08), GO:0005840 (ribosome; P = 1.40E−07), and GO:0015935 (small ribosomal subunit; P = 0.019). The genes in these categories coded for both large and small ribosomal subunits (see Table S2 in the supplemental material) and were 10- to 42-fold more abundant in T. gondii sporulated oocysts than in H. hammondi. In addition to GO categories, we also analyzed all other currently available ToxoDB gene annotations using GeneMerge. We found that the Tg-high and Hh-high sets were not significantly enriched for genes in any particular annotation except the GO categories listed above, including the presence or absence of a putative signal peptide.
FIG 6.
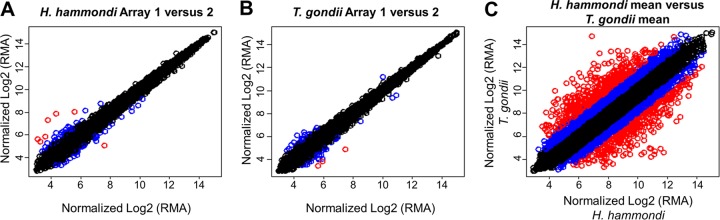
Plots of normalized, log2-transformed hybridization intensities within (A and B) and between (C) species. The plots in panels A and B are derived from replicate arrays within each species, and the plot in panel C is the mean across the 3 arrays for each species. The intensities for each gene are color-coded based on the absolute values of the fold difference in transcript abundance as follows: black, ≤2-fold; blue, between 2- and 4-fold; red, ≥4-fold. RMA, robust multiarray average.
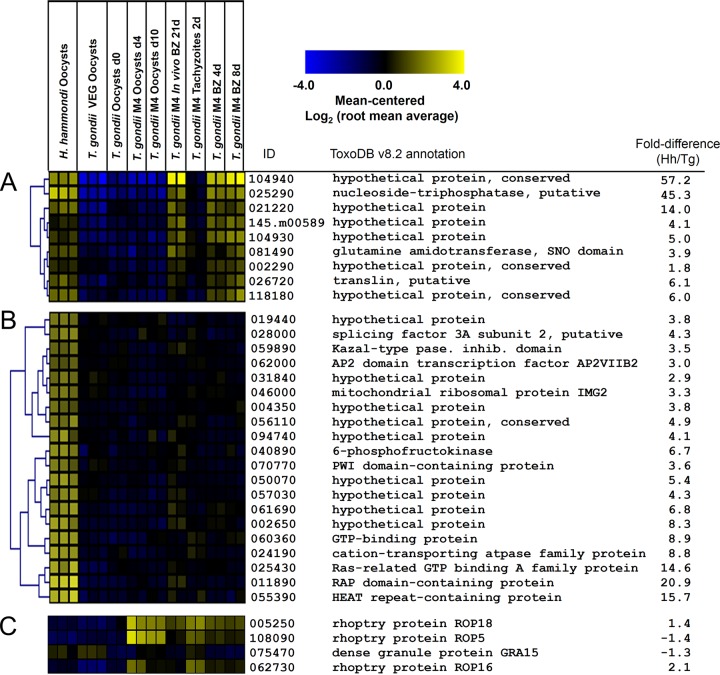
We also compared these transcript data to the previously published transcriptional profile from multiple life stages of T. gondii type II strain M4 (including unsporulated and sporulated oocysts, tachyzoites, and in vitro and in vivo bradyzoite cysts [39, 40]). Overall, the majority of the Tg-high set were expressed not only in T. gondii oocyst/sporozoite stages but also in other M4 life stages. This was in contrast to the Hh-high gene set, and by comparing these data to the M4 data set, we identified two distinct clusters for further analysis: one where Hh-high transcripts were low in all T. gondii VEG and M4 life stages analyzed (“Hh-specific”) and another where Hh-high transcripts were high in T. gondii cyst stages (“Hh-TgCyst-high”; see Fig. S5). In support of the existence of an Hh-TgCyst-high set was a significant (P = 0.0011) overlap between the Hh-high gene set and an empirically determined gene set that is upregulated during the transition from tachyzoites to bradyzoites in vivo and/or in vitro in T. gondii strain M4. To identify these genes in an unbiased way, we used Pavlidis template matching (47) at an adjusted P value threshold of 1 × 10−6 and identified 9 Hh-TgCyst-high genes and 38 Hh-specific genes (Fig. 7A and B, respectively) that significantly matched the respective templates. GO analysis on both gene sets did not identify any significant GO category enrichment. However, included in the Hh-specific set was a predicted AP2 transcription factor, APVIIB2. Transcript abundance for this gene was low in T. gondii VEG sporulated oocysts and all M4 life cycle stages (Fig. 7B). While this was the only AP2 transcription factor family member that was of significantly higher abundance in H. hammondi than in T. gondii, among the 53 AP2 family members analyzed on the microarray, APIX8 and APVIII5 transcript levels were also 6-fold higher in H. hammondi than in T. gondii VEG sporulated oocysts (see Fig. S9 in the supplemental material), and these transcripts are upregulated during the tachyzoite-to-bradyzoite transition in T. gondii strain M4 (arrowheads in Fig. S9). Genes in the Hh-TgCyst-high set included genes for a bradyzoite-specific NTPase and multiple hypothetical proteins (Fig. 7B). In contrast to these major differences in transcription between T. gondii and H. hammondi, we detected no significant expression differences between H. hammondi and T. gondii VEG sporulated oocyst transcript levels for T. gondii secreted effectors like ROP5, ROP18, ROP16, and GRA15 (Fig. 7C). Overall, these data show that H. hammondi sporulated oocysts have a unique expression profile compared to that of T. gondii sporulated oocysts, and some of these differentially regulated genes are highly expressed in T. gondii M4 bradyzoites.
FIG 7.
Transcript abundance in H. hammondi HhCatGer041 and T. gondii VEG sporozoites for 9 genes of higher transcript abundance in H. hammondi sporozoites as well as T. gondii cyst stages (Hh-TgCyst-high) (A), a subset of genes of high abundance in H. hammondi sporozoites that were of low abundance in T. gondii VEG and all M4 life cycle stages (B), and known T. gondii effectors (rhoptry proteins ROP5, ROP16, and ROP18 and dense granule protein GRA15). Data from a previously published data set profiling the transcriptome of multiple life cycle stages of T. gondii strain M4 (GEO series GSE32427) are shown for comparison, and gene identifications are based on ME49B7 annotation version 7.2.
DISCUSSION
Our recently published genome sequence of an isolate of H. hammondi (HhCatGer041 [28]; GenBank accession number AVCM00000000.1) revealed a high degree of genomic synteny and conservation with T. gondii, further confirming H. hammondi as the closest known extant relative of T. gondii (28). Despite this high sequence similarity, there are dramatic phenotypic differences between H. hammondi and T. gondii. Unlike T. gondii, H. hammondi spontaneously converts to slow-growing tissue cyst stages in vitro and cannot be propagated indefinitely in vitro (9, 16). This has also been shown in vitro for another Hammondia sp. isolated from foxes (57), now named H. triffittae (58). H. hammondi is relatively avirulent in mice (including interferon gamma knockouts [9, 16]) and has never been shown to cause disease in humans. The genetics underlying these phenotypic differences are unknown. They could be due to differences in gene content due to gene gain and loss during and after divergence of these species. The also could be due to distinct transcriptional profiles. Building off previous work showing that H. hammondi orthologs of the T. gondii virulence effectors ROP18 and ROP5 are functionally conserved when expressed in T. gondii (28), in the present study we examined the efficacy of the syntenic H. hammondi orthologs of GRA15 (27) and ROP16 (25). Our assay for functional conservation was 2-fold: (i) was the H. hammondi ortholog effectively expressed in T. gondii, and (ii) could the H. hammondi ortholog complement a known cell signaling phenotype?
The H. hammondi GRA15 promoter was functional when expressed heterologously in T. gondii, while the H. hammondi ROP16 promoter was far less effective (see Table S1 in the supplemental material). The promoters for H. hammondi ROP16 and T. gondii ROP16 are highly conserved, but there is a16-bp deletion just upstream of the putative transcriptional start site in the H. hammondi ROP16 promoter. No such differences exist in the H. hammondi and T. gondii GRA15 upstream sequences. The deletion in the HhROP16 promoter is similar to previous observations for the H. hammondi and T. gondii ROP18 promoters. In the ROP18 gene there is a 107-bp sequence present in T. gondii strains that actively express ROP18 (such as strain types I and II [15, 23]) as well as HhROP18 (28) but absent in T. gondii strains that do not express ROP18 (such as members of the type III lineage [15, 23]). The HhROP18 promoter is fully active in luciferase reporter assays and drives HhROP18 protein expression in T. gondii, and this dramatically alters virulence; this 107-bp sequence is both necessary and sufficient for ROP18 promoter-driven reporter expression (28). In the present study, we found that the 16 bp deleted in the HhROP16 promoter play an important role in the ability of the ROP16 upstream sequence to drive reporter gene expression, suggesting that this is an important core promoter sequence for ROP16. This result further confirms the utility of comparative genomic analysis to identify regulatory regions that are important for the expression of T. gondii effector proteins like ROP18 and ROP16. It is important to note that the lack of activity of the H. hammondi ROP16 promoter when expressed in T. gondii does not prove that the H. hammondi ROP16 gene is not expressed in its native context (i.e., within H. hammondi itself). At least in sporulated oocysts there were no differences in ROP16 transcript abundance between H. hammondi and T. gondii VEG sporozoites, although the ROP16 transcript was detectable in both species. Transcriptome analyses of H. hammondi tachyzoites will be necessary to determine if this difference in promoter function in a heterologous expression system is recapitulated in vivo. While such experiments are significantly hampered by the inability to grow H. hammondi indefinitely in vitro, deep RNA sequencing technology will be useful to circumvent this problem.
When expressed in a type I strain of T. gondii, H. hammondi GRA15 leads to significantly more NF-κB p65 activation than a wild-type type I strain. It has been shown that the type II allele of T. gondii GRA15 is a much more potent activator of NF-κB signaling than the alleles from T. gondii clonotypes I and III (27). H. hammondi GRA15 can increase NF-κB p65 activation in a T. gondii type I strain to levels that are very similar to that of a wild-type T. gondii type II strain (Fig. 2B), suggesting that H. hammondi GRA15 behaves like a T. gondii type II GRA15 allele. Our sequence analysis shows that while overall HhGRA15 is most similar to T. gondii GRA15 alleles from types I and III, this is quantitatively driven by the fact that HhGRA15 shares an 84-amino-acid sequence with TgGRA15I/III that is deleted in TgGRA15II. These data indicate that the sequence in HhGRA15 and TgGRA15I/III relative to TgGRA15II is ancestral to the T. gondii lineage (and therefore is likely to be a deletion in TgGRA15II). Moreover, the fact that expressing HhGRA15 in T. gondii type I increases its ability to activate NF-κB to levels that are similar to those in type II strains indicates that the C-terminal deletion in TgGRA15II is not responsible for differences in the ability of T. gondii alleles to activate NF-κB, a possibility proposed by Rosowski et al. (27). Further experiments, such as complementing a type I strain with a type I GRA15 allele and comparing its efficacy to those of HhGRA15 and TgGRA15II, will be necessary to determine whether HhGRA15 behaves identically to TgGRA15II. If it does, this would implicate only 3 polymorphisms present in both TgGRA15II and HhGRA15 as being potentially responsible for the significant phenotypic differences between TgGRA15II and TgGRA15I/III.
When expressed off a highly active promoter in T. gondii, HhROP16 is less efficient at trafficking to the host cell nucleus than TgROP16I, but it still increases STAT6 nuclear translocation when expressed in a T. gondii type II strain. At the amino acid level, H. hammondi ROP16 is most similar to T. gondii type II ROP16, and therefore, the increase in STAT6 activation shown in Fig. 4D could be due to gene dosage effects (T. gondii type II:HhROP16 also has an endogenous copy of TgROP16) rather than HhROP16 being more functionally similar to TgROP16I/III. To test this, we created a type II T. gondii parasite clone with an additional copy of ROP16II (driven by the same TgGRA1 promoter as HhROP16) and compared STAT6 activation driven by this strain to those in wild-type type II T. gondii and type II T. gondii expressing HhROP16. We found that T. gondii type II:TgROP16II was no more capable of STAT6 activation than T. gondii type II:WT, while again T. gondii type II:HhROP16 significantly activated STAT6 (Fig. 4E). These data further support a major role for residue 503 in HhROP16 and TgROP16I/III, which is a serine in both HhROP16 and TgROP16I/III and has been shown to be required for the phosphorylation of STAT3 by TgROP16I/III (and which is a serine in TgROP16II which does not phosphorylate STAT3 [25, 54]).
As mentioned above, these data do not address whether the promoters for H. hammondi ROP16 and GRA15 are actually functional in their native context within H. hammondi during acute infection. Such an analysis is hampered by the fact that H. hammondi sporozoites spontaneously convert to bradyzoite cysts in vitro and to date cannot be subcultured (5, 9, 16, 59). This is in contrast even to T. gondii VEG strain sporozoites, which, although they also form bradyzoite cysts spontaneously in vitro (60), can be subcultured and eventually grow as rapidly growing tachyzoites (60).
The transcriptional data in the present study are from sporozoites isolated from T. gondii and H. hammondi oocysts and represent the first transcriptional comparison between these species. These data show that the sporulated oocyst transcriptomes for these species are highly distinct. Among the genes of greater abundance in H. hammondi than in T. gondii VEG, a subset were highly expressed only in H. hammondi sporozoites compared to both T. gondii VEG and M4 sporozoites and multiple M4 life cycle stages. These “Hh-specific” genes are interesting candidates that may underlie phenotypic differences between these species. Of particular interest in the Hh-specific gene set was the AP2 transcription factor AP2VIIB2 (Fig. 7B), which not only is of low abundance in the M4 data set but also is consistently of low abundance (<40th percentile) in 15 microarray and transcriptome sequencing expression data sets from multiple strains and life stages available for this gene on ToxoDB (v8.2 gene name, TGME49_262000). Given emerging discoveries regarding the importance of the AP2 transcription factor family in T. gondii developmental processes, particularly those involving tachyzoite-to-bradyzoite conversion (61–63)), this is a particularly intriguing candidate gene for mediating transcriptional differences between these species, at least in sporulated oocysts.
Another subset of Hh-high transcripts are developmentally regulated during the tachyzoite-to-bradyzoite transition in T. gondii strain M4 (both in vitro and in vivo). Two other AP2 family members were over 6-fold more abundant in H. hammondi sporulated oocysts than in T. gondii, and while these were not significantly different, they do appear to also be upregulated in strain M4 bradyzoites (both in vivo and in vitro). In addition, multiple members of the Hh-high gene set are at least slightly upregulated in M4 bradyzoites compared to tachyzoites. This observation, coupled with their significantly higher expression in H. hammondi sporulated oocysts than in T. gondii VEG, fits with the dramatic growth differences between these species both in vitro and in vivo. The expression patterns of “bradyzoite” genes in H. hammondi sporulated oocysts may be a reflection of their place on the developmental spectrum between tachyzoite and bradyzoite. This spectrum has been described previously for T. gondii strains that differ in their growth characteristics in vitro (e.g., VEG compared to RH [64]) but has yet to be described in multispecies comparisons like those outlined in the present study.
Transcriptional comparisons between T. gondii and Neospora caninum, a fellow coccidian that is more diverged from T. gondii than H. hammondi, have been reported (65). N. caninum tachyzoites have significantly lower ROP16 transcript levels than T. gondii, and it was proposed that despite the conserved leucine residue crucial for ROP16-driven STAT3 phosphorylation in N. caninum ROP16, this strain of N. caninum would be unlikely to modulate the STAT3 (or possibly STAT6) pathway (65). This has yet to be tested empirically, and attempts to align the upstream regions of NcROP16, HhROP16, and TgROP16 were not possible given the high divergence of the NcROP16 upstream sequence.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge John Boothroyd (Stanford University) for providing the ROP16HA-complemented T. gondii strain and Eric Polinko (University of Pittsburgh) for computing support.
This work was supported by a Pew Scholarship in the Biomedical Sciences to J.P.B. and by University of Pittsburgh Biological Sciences summer and academic year fellowships funded by the Howard Hughes Medical Institute to K.A.W., R.A.D., A.L.B., and A.R.S.
We declare no conflict of interest.
Footnotes
Published ahead of print 3 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00215-14.
REFERENCES
- 1.Jones JL, Kruszon-Moran D, Wilson M, McQuillan G, Navin T, McAuley JB. 2001. Toxoplasma gondii infection in the United States: seroprevalence and risk factors. Am. J. Epidemiol. 154:357–365. 10.1093/aje/154.4.357. [DOI] [PubMed] [Google Scholar]
- 2.Israelski DM, Remington JS. 1993. Toxoplasmosis in patients with cancer. Clin. Infect. Dis. 17(Suppl 2):S423–S435. [DOI] [PubMed] [Google Scholar]
- 3.Luft BJ, Hafner R, Korzun AH, Leport C, Antoniskis D, Bosler EM, Bourland DD, III, Uttamchandani R, Fuhrer J, Jacobson J, Morlat P, Vilde J-L, Remington JS, ACTG 077p/ANRS 009 Study Team 1993. Toxoplasmic encephalitis in patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 329:995–1000. [DOI] [PubMed] [Google Scholar]
- 4.Remington JS, Klein JO. 2001. Infectious diseases of the fetus and newborn infant, 5th ed. Saunders, Philadelphia, PA. [Google Scholar]
- 5.Frenkel JK, Dubey JP. 1975. Hammondia hammondi gen. nov., sp. nov., from domestic cats, a new coccidian related to Toxoplasma and Sarcocystis. Z. Parasitenkd. 46:3–12. 10.1007/BF00383662. [DOI] [PubMed] [Google Scholar]
- 6.Carme B, Demar M, Ajzenberg D, Darde ML. 2009. Severe acquired toxoplasmosis caused by wild cycle of Toxoplasma gondii, French Guiana. Emerg. Infect. Dis. 15:656–658. 10.3201/eid1504.081306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubey JP. 1998. Advances in the life cycle of Toxoplasma gondii. Int. J. Parasitol. 28:1019–1024. 10.1016/S0020-7519(98)00023-X. [DOI] [PubMed] [Google Scholar]
- 8.Wong SY, Remington JS. 1993. Biology of Toxoplasma gondii. AIDS 7:299–316. [DOI] [PubMed] [Google Scholar]
- 9.Dubey JP, Sreekumar C. 2003. Redescription of Hammondia hammondi and its differentiation from Toxoplasma gondii. Int. J. Parasitol. 33:1437–1453. 10.1016/S0020-7519(03)00141-3. [DOI] [PubMed] [Google Scholar]
- 10.Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM. 1999. Phylogenetic analysis based on full-length large subunit ribosomal RNA gene sequence comparison reveals that Neospora caninum is more closely related to Hammondia heydorni than to Toxoplasma gondii. Int. J. Parasitol. 29:1545–1556. 10.1016/S0020-7519(99)00150-2. [DOI] [PubMed] [Google Scholar]
- 11.Su C, Evans D, Cole RH, Kissinger JC, Ajioka JW, Sibley LD. 2003. Recent expansion of Toxoplasma through enhanced oral transmission. Science 299:414–416. 10.1126/science.1078035. [DOI] [PubMed] [Google Scholar]
- 12.Su C, Khan A, Zhou P, Majumdar D, Ajzenberg D, Darde ML, Zhu XQ, Ajioka JW, Rosenthal BM, Dubey JP, Sibley LD. 2012. Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc. Natl. Acad. Sci. U. S. A. 109:5844–5849. 10.1073/pnas.1203190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan A, Miller N, Roos DS, Dubey JP, Ajzenberg D, Darde ML, Ajioka JW, Rosenthal B, Sibley LD. 2011. A monomorphic haplotype of chromosome Ia is associated with widespread success in clonal and nonclonal populations of Toxoplasma gondii. mBio 2(6):e00228–11. 10.1128/mBio.00228-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle JP, Saeij JP, Boothroyd JC. 2007. Toxoplasma gondii: inconsistent dissemination patterns following oral infection in mice. Exp. Parasitol. 116:302–305. 10.1016/j.exppara.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Saeij JP, Boyle JP, Coller S, Taylor S, Sibley LD, Brooke-Powell ET, Ajioka JW, Boothroyd JC. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314:1780–1783. 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubey JP, Tilahun G, Boyle JP, Schares G, Verma SK, Ferreira LR, Oliveira S, Tiao N, Darrington C, Gebreyes WA. 2013. Molecular and biological characterization of first isolates of Hammondia hammondi from cats from Ethiopia. J. Parasitol. 99:614–618. 10.1645/12-51.1. [DOI] [PubMed] [Google Scholar]
- 17.Araujo FG, Dubey JP, Remington JS. 1984. Antigenic similarity between the coccidian parasites Toxoplasma gondii and Hammondia hammondi. J. Protozool. 31:145–147. 10.1111/j.1550-7408.1984.tb04304.x. [DOI] [PubMed] [Google Scholar]
- 18.Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. 2011. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc. Natl. Acad. Sci. U. S. A. 108:9631–9636. 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckenstein MC, Reese ML, Konen-Waisman S, Boothroyd JC, Howard JC, Steinfeldt T. 2012. A Toxoplasma gondii pseudokinase inhibits host IRG resistance proteins. PLoS Biol. 10:e1001358. 10.1371/journal.pbio.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khaminets A, Hunn JP, Konen-Waisman S, Zhao YO, Preukschat D, Coers J, Boyle JP, Ong YC, Boothroyd JC, Reichmann G, Howard JC. 2010. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell. Microbiol. 12:939–961. 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niedelman W, Gold DA, Rosowski EE, Sprokholt JK, Lim D, Farid Arenas A, Melo MB, Spooner E, Yaffe MB, Saeij JP. 2012. The rhoptry proteins ROP18 and ROP5 mediate Toxoplasma gondii evasion of the murine, but not the human, interferon-gamma response. PLoS Pathog. 8:e1002784. 10.1371/journal.ppat.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reese ML, Zeiner GM, Saeij JP, Boothroyd JC, Boyle JP. 2011. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc. Natl. Acad. Sci. U. S. A. 108:9625–9630. 10.1073/pnas.1015980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor S, Barragan A, Su C, Fux B, Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, White M, Wootton JC, Sibley LD. 2006. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 314:1776–1780. 10.1126/science.1133643. [DOI] [PubMed] [Google Scholar]
- 24.Butcher BA, Fox BA, Rommereim LM, Kim SG, Maurer KJ, Yarovinsky F, Herbert DR, Bzik DJ, Denkers EY. 2011. Toxoplasma gondii rhoptry kinase ROP16 activates STAT3 and STAT6 resulting in cytokine inhibition and arginase-1-dependent growth control. PLoS Pathog. 7:e1002236. 10.1371/journal.ppat.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeij JP, Coller S, Boyle JP, Jerome ME, White MW, Boothroyd JC. 2007. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature 445:324–327. 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen KD, Wang Y, Wojno ED, Shastri AJ, Hu K, Cornel L, Boedec E, Ong YC, Chien YH, Hunter CA, Boothroyd JC, Saeij JP. 2011. Toxoplasma polymorphic effectors determine macrophage polarization and intestinal inflammation. Cell Host Microbe 9:472–483. 10.1016/j.chom.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosowski EE, Lu D, Julien L, Rodda L, Gaiser RA, Jensen KD, Saeij JP. 2011. Strain-specific activation of the NF-kappaB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 208:195–212. 10.1084/jem.20100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walzer KA, Adomako-Ankomah Y, Dam RA, Herrmann DC, Schares G, Dubey JP, Boyle JP. 2013. Hammondia hammondi, an avirulent relative of Toxoplasma gondii, has functional orthologs of known T. gondii virulence genes. Proc. Natl. Acad. Sci. U. S. A. 110:7446–7451. 10.1073/pnas.1304322110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakaguri H, Suzuki Y, Katayama T, Kawashima S, Kibukawa E, Hiranuka K, Sasaki M, Sugano S, Watanabe J. 2009. Full-Malaria/Parasites and Full-Arthropods: databases of full-length cDNAs of parasites and arthropods, update 2009. Nucleic Acids Res. 37:D520–D525. 10.1093/nar/gkn856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trujillo M, Donald RG, Roos DS, Greene PJ, Santi DV. 1996. Heterologous expression and characterization of the bifunctional dihydrofolate reductase-thymidylate synthase enzyme of Toxoplasma gondii. Biochemistry 35:6366–6374. 10.1021/bi952923q. [DOI] [PubMed] [Google Scholar]
- 31.Hartley JL, Temple GF, Brasch MA. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10:1788–1795. 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle JP, Saeij JP, Harada SY, Ajioka JW, Boothroyd JC. 2008. Expression quantitative trait locus mapping of Toxoplasma genes reveals multiple mechanisms for strain-specific differences in gene expression. Eukaryot. Cell 7:1403–1414. 10.1128/EC.00073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. 2007. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 3:e77. 10.1371/journal.ppat.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donald RG, Carter D, Ullman B, Roos DS. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019. [DOI] [PubMed] [Google Scholar]
- 35.Parmley SF, Sgarlato GD, Remington JS. 1993. Genomic and corrected cDNA sequence of the P28 gene from Toxoplasma gondii. Mol. Biochem. Parasitol. 57:161–165. 10.1016/0166-6851(93)90253-T. [DOI] [PubMed] [Google Scholar]
- 36.Rome ME, Beck JR, Turetzky JM, Webster P, Bradley PJ. 2008. Intervacuolar transport and unique topology of GRA14, a novel dense granule protein in Toxoplasma gondii. Infect. Immun. 76:4865–4875. 10.1128/IAI.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahl A, Davis PH, Behnke M, Dzierszinski F, Jagalur M, Chen F, Shanmugam D, White MW, Kulp D, Roos DS. 2010. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics 11:603. 10.1186/1471-2164-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fritz HM, Buchholz KR, Chen X, Durbin-Johnson B, Rocke DM, Conrad PA, Boothroyd JC. 2012. Transcriptomic analysis of Toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PLoS One 7:e29998. 10.1371/journal.pone.0029998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchholz KR, Fritz HM, Chen X, Durbin-Johnson B, Rocke DM, Ferguson DJ, Conrad PA, Boothroyd JC. 2011. Identification of tissue cyst wall components by transcriptome analysis of in vivo and in vitro Toxoplasma gondii bradyzoites. Eukaryot. Cell 10:1637–1647. 10.1128/EC.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Development Core Team. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 42.Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 43.Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573:83–92. 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 44.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. 2006. TM4 microarray software suite. Methods Enzymol. 411:134–193. 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 45.Castillo-Davis CI, Hartl DL. 2003. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19:891–892. 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 46.Kamau E, Meehan T, Lavine MD, Arrizabalaga G, Mustata Wilson G, Boyle J. 2011. A novel benzodioxole-containing inhibitor of Toxoplasma gondii growth alters the parasite cell cycle. Antimicrob. Agents Chemother. 55:5438–5451. 10.1128/AAC.00455-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pavlidis P, Noble WS. 2001. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2:RESEARCH0042. 10.1186/gb-2001-2-10-research0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grigg ME, Ganatra J, Boothroyd JC, Margolis TP. 2001. Unusual abundance of atypical strains associated with human ocular toxoplasmosis. J. Infect. Dis. 184:633–639. 10.1086/322800. [DOI] [PubMed] [Google Scholar]
- 49.Sibley LD, Boothroyd JC. 1992. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature 359:82–85. 10.1038/359082a0. [DOI] [PubMed] [Google Scholar]
- 50.Khan A, Dubey JP, Su C, Ajioka JW, Rosenthal BM, Sibley LD. 2011. Genetic analyses of atypical Toxoplasma gondii strains reveal a fourth clonal lineage in North America. Int. J. Parasitol. 41:645–655. 10.1016/j.ijpara.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saeij JP, Boyle JP, Boothroyd JC. 2005. Differences among the three major strains of Toxoplasma gondii and their specific interactions with the infected host. Trends Parasitol. 21:476–481. 10.1016/j.pt.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 52.Boyle JP, Rajasekar B, Saeij JP, Ajioka JW, Berriman M, Paulsen I, Roos DS, Sibley LD, White MW, Boothroyd JC. 2006. Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 103:10514–10519. 10.1073/pnas.0510319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ong YC, Reese ML, Boothroyd JC. 2010. Toxoplasma rhoptry protein 16 (ROP16) subverts host function by direct tyrosine phosphorylation of STAT6. J. Biol. Chem. 285:28731–28740. 10.1074/jbc.M110.112359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto M, Standley DM, Takashima S, Saiga H, Okuyama M, Kayama H, Kubo E, Ito H, Takaura M, Matsuda T, Soldati-Favre D, Takeda K. 2009. A single polymorphic amino acid on Toxoplasma gondii kinase ROP16 determines the direct and strain-specific activation of Stat3. J. Exp. Med. 206:2747–2760. 10.1084/jem.20091703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sibley LD, Niesman IR, Parmley SF, Cesbron-Delauw MF. 1995. Regulated secretion of multi-lamellar vesicles leads to formation of a tubulo-vesicular network in host-cell vacuoles occupied by Toxoplasma gondii. J. Cell Sci. 108(Part 4):1669–1677. [DOI] [PubMed] [Google Scholar]
- 56.Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. 2009. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 284:478–485. 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 57.Schares G, Meyer J, Barwald A, Conraths FJ, Riebe R, Bohne W, Rohn K, Peters M. 2003. A Hammondia-like parasite from the European fox (Vulpes vulpes) forms biologically viable tissue cysts in cell culture. Int. J. Parasitol. 33:229–234. 10.1016/S0020-7519(03)00009-2. [DOI] [PubMed] [Google Scholar]
- 58.Gjerde B, Dahlgren SS. 2011. Hammondia triffittae n. comb. of foxes (Vulpes spp.): biological and molecular characteristics and differentiation from Hammondia heydorni of dogs. Parasitology 138:303–321. 10.1017/S0031182010001265. [DOI] [PubMed] [Google Scholar]
- 59.Frenkel JK, Dubey JP. 1975. Hammondia hammondi: a new coccidium of cats producing cysts in muscle of other mammals. Science 189:222–224. 10.1126/science.806116. [DOI] [PubMed] [Google Scholar]
- 60.Jerome ME, Radke JR, Bohne W, Roos DS, White MW. 1998. Toxoplasma gondii bradyzoites form spontaneously during sporozoite-initiated development. Infect. Immun. 66:4838–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radke JB, Lucas O, De Silva EK, Ma Y, Sullivan WJ, Jr, Weiss LM, Llinas M, White MW. 2013. ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst. Proc. Natl. Acad. Sci. U. S. A. 110:6871–6876. 10.1073/pnas.1300059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, Sibley LD, White MW. 2010. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One 5:e12354. 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker R, Gissot M, Croken MM, Huot L, Hot D, Kim K, Tomavo S. 2013. The Toxoplasma nuclear factor TgAP2XI-4 controls bradyzoite gene expression and cyst formation. Mol. Microbiol. 87:641–655. 10.1111/mmi.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, White MW. 2005. The transcriptome of Toxoplasma gondii. BMC Biol. 3:26. 10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reid AJ, Vermont SJ, Cotton JA, Harris D, Hill-Cawthorne GA, Konen-Waisman S, Latham SM, Mourier T, Norton R, Quail MA, Sanders M, Shanmugam D, Sohal A, Wasmuth JD, Brunk B, Grigg ME, Howard JC, Parkinson J, Roos DS, Trees AJ, Berriman M, Pain A, Wastling JM. 2012. Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: coccidia differing in host range and transmission strategy. PLoS Pathog. 8:e1002567. 10.1371/journal.ppat.1002567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.