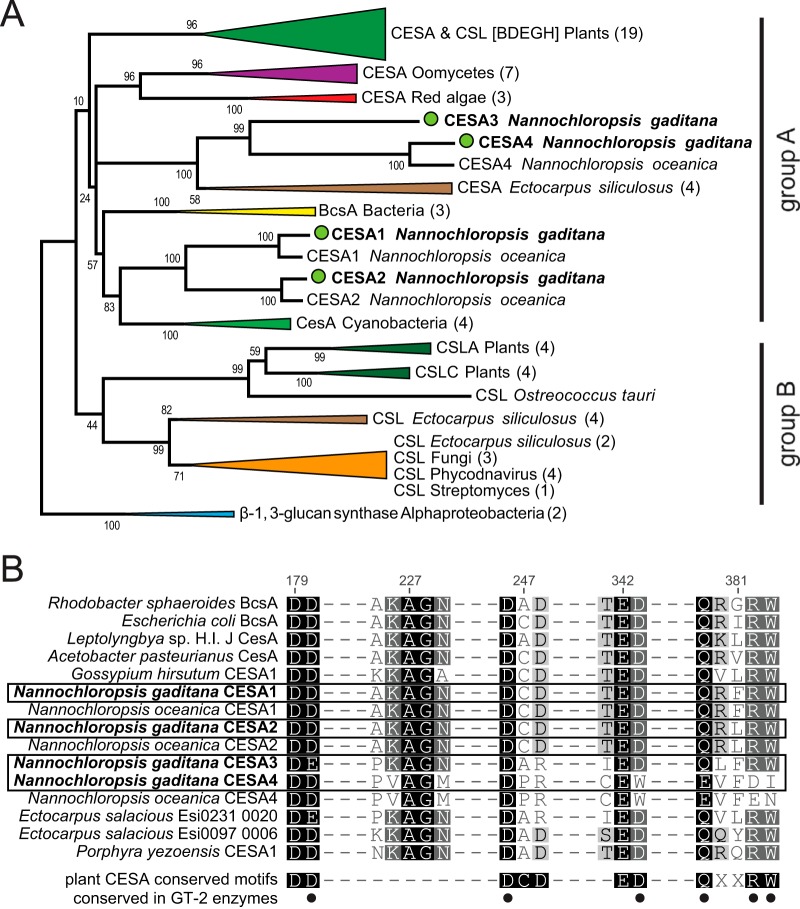
FIG 1.
(A) Phylogenetic tree of cellulose synthases (CESA) and cellulose synthase-like proteins (CSL). The tree was constructed using the maximum-likelihood (ML) approach. N. gaditana sequences are marked with green circles. Numbers indicate bootstrap values in the ML analysis. Numbers in parentheses indicate the numbers of sequences compressed in subtrees. Groups A and B are described in the text. The full list of accession numbers from the proteins used can be found in Table S1 in the supplemental material. (B) Multiple sequence alignments of the motifs common to cellulose synthases. Numbers indicate the amino acid positions in the Rhodobacter sphaeroides BcsA protein (58). Dashes do not represent gaps in the alignment, and not all aligned amino acids are shown. Dots indicate the conserved residues (D, D, D, and QXXRW) found in characterized cellulose synthases within the glycosyl transferase 2 (GT-2) family, as cataloged by the Carbohydrate-Active enZymes (CAZy) database.