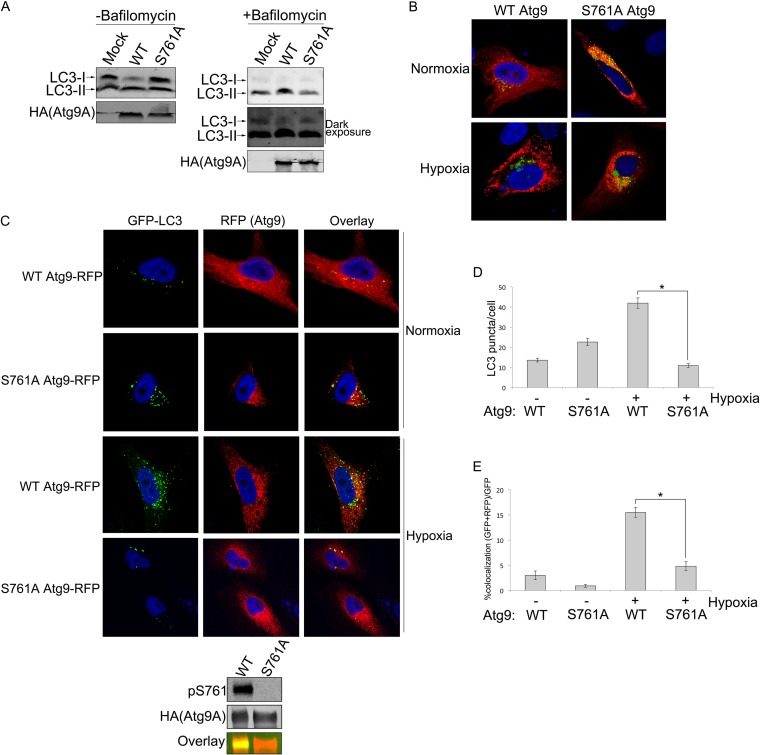
FIG 6.
Phosphorylation of S761 is required for autophagy induction and Atg9A localization to LC3-positive autophagosomes in hypoxia. (A) HEK-293 cells expressing WT Atg9A or the S761A mutant or that were mock transfected were treated with hypoxia for 12 h with or without 100 nM bafilomycin A1. Cells were lysed and immunoblotted for endogenous LC3-I and LC3-II. The intensity of LC3 bands was quantified by Li-Cor infrared imaging in order to determine the LC3-II/LC3-I ratio. The combined results from five separate experiments are shown in the bar graph with means ± standard errors. (B) U2OS cells infected with Golgi-GFP Bac Mam (Life Technologies, Inc.) were transfected with the RFP-tagged Atg9A WT or S761A mutant and treated with hypoxia or normoxia for 12 h. After fixing, cells were analyzed by confocal microscopy. (C) HeLa cells stably expressing GFP-LC3 were transfected with the RFP-tagged Atg9A WT or S761A mutant and treated with hypoxia or normoxia for 12 h. After fixing, cells were analyzed by using confocal microscopy. The panel below the images shows phosphorylation at S761 in hypoxia-treated (12 h) HeLa cells. (D) LC3 punctate structures were counted in RFP-tagged Atg9A WT- or S761A mutant-expressing cells. Means ± standard errors are shown. The statistical significance of the differences between the WT and the S761A mutant was calculated by using two-tailed unpaired Student's t test with equal variance (n = 86 [WT normoxia], n = 76 [S761A mutant normoxia], n = 84 [WT hypoxia], and n = 87 [S761A mutant hypoxia]). *, P value of 1.43E−22. (E) Percentage of GFP-LC3 punctate structures colocalizing with RFP-Atg9A (WT or S761A mutant) determined by manually counting punctate structures in multiple fields (the number of GFP-LC3 and RFP-Atg9A puncta divided by the total number of GFP-LC3 puncta). Means ± standard errors are shown. The statistical significance of the difference between the WT and the S761A mutant was calculated by using two-tailed unpaired Student's t test with equal variance (n = 86 [WT normoxia], n = 76 [S761A mutant normoxia], n = 84 [WT hypoxia], and n = 87 [S761A mutant hypoxia]). *, P value of 3.87E−13.