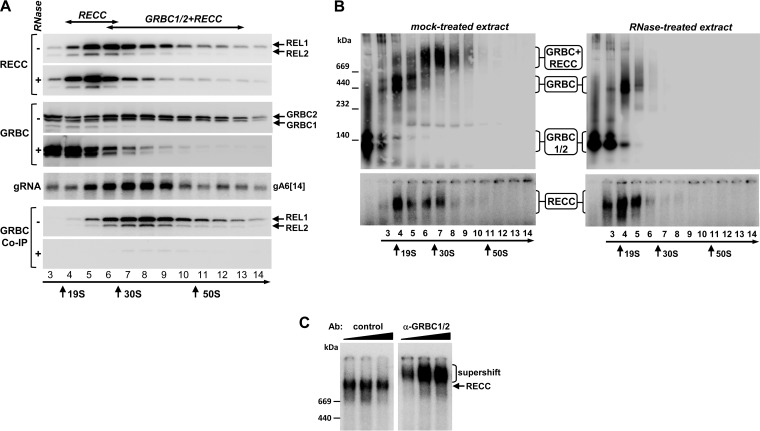
FIG 1.
Cocomplex interactions between editing core and gRNA binding complexes. (A) The impact of RNase treatment on RECC and GRBC1/2 cocomplex interactions. Mock-treated extracts (−) and extracts preincubated with RNase A (0.1 mg/ml) and RNase T1 (1,000 U/ml) (+) were separated on 10% to 30% glycerol gradients for 4 h at 178,000 × g. Sedimentation patterns of the RNA editing core complex were visualized by self-adenylation of RNA editing ligases REL1 and REL2 in the presence of [α-32P]ATP (58). Following immunoprecipitation, anti-GRBC1/2 antibody-coated beads were incubated with [α-32P]ATP to label REL1 and REL2 RNA ligases as a means of detecting the editing core complex. Bound proteins were eluted with SDS gel loading buffer. Alternatively, RNA was extracted from each fraction and separated on a 10% denaturing polyacrylamide gel (PAGE) for hybridization with a gRNA A6[14]-specific probe. (B) RNase treatment eliminates the ∼40S particle. The gradient fractions described for panel A were separated on native 3% to 12% PAGE to estimate the molecular mass of untreated and RNase-resistant RECC and GRBC1/2-containing complexes. (C) “Supershift” of RECC with α-GRBC1/2 antibody. Preadenylated fraction 8 was incubated with 0.1, 0.3, and 1 μg of antigen-purified control and α-GRBC1/2 antibodies and separated on 3% to 12% native PAGE.