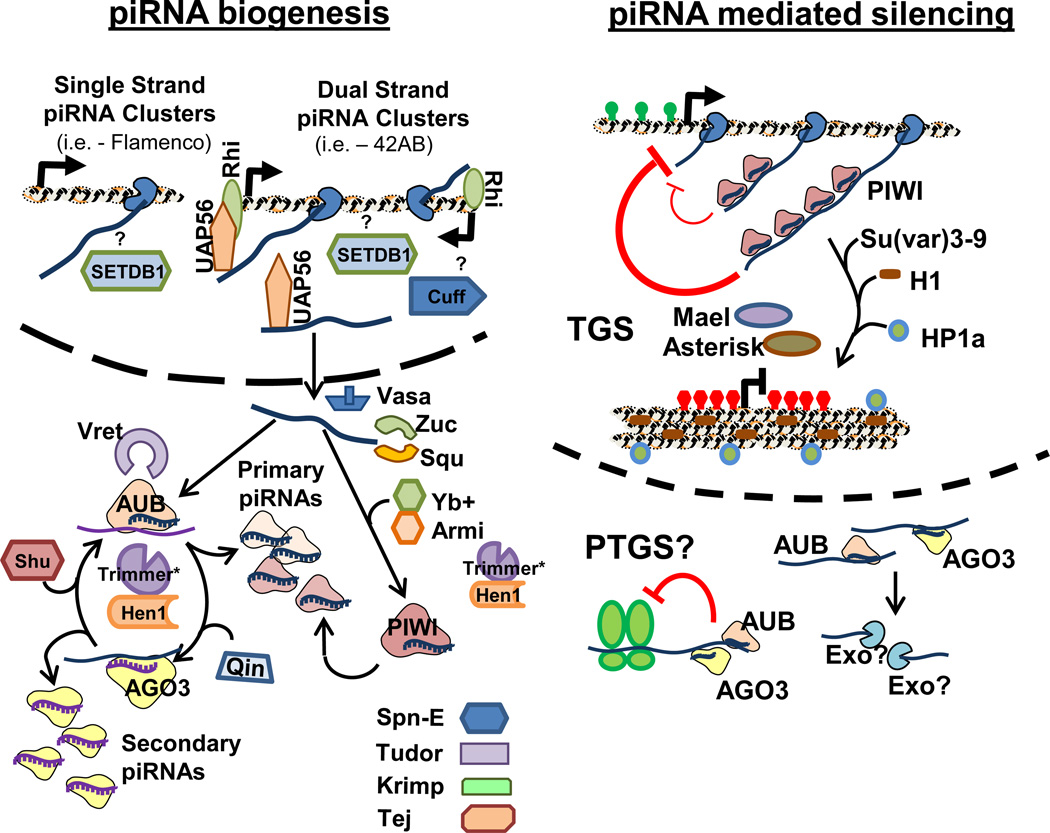
Figure 3. Current models of piRNA biogenesis and piRNA mediated gene silencing mechanisms in Drosophila.
Left) Depiction of known roles for factors genetically implicated in Drosophila piRNA biogenesis. More factors are known to regulate dual strand than single strand piRNA clusters. The piRNA precursor transcripts either are directly processed into primary piRNAs bound by AUB and PIWI, or engage in a subsequent secondary piRNA “ping-pong” amplification by AUB and AGO3. Right) Since AUB and AGO3 are in perinuclear nuage and cytoplasm; they may mediate post-transcriptional gene silencing. The nuclear PIWI binds to nascent transposon transcripts, where it can recruit chromatin modifying factors to induce a repressive chromatin state. Many other factors not shown in this diagram that may affect TE silencing have been identified in knockdown screens, but placement into the model is unclear, as is the role for tudor-domain containing factors like Spn-E, Tudor, Krimp, and Tej.