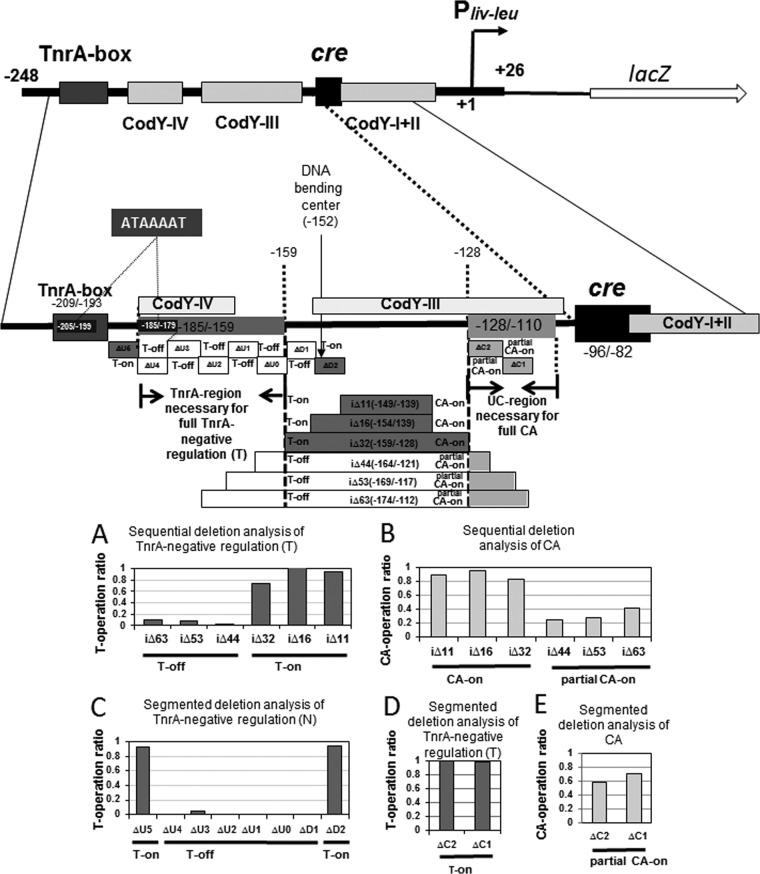
FIG 7.
Sequential and segmented deletion analysis of the regions necessary for full CA and TnrA-negative regulation between the TnrA box and cre site. The cre and CodY sites and the TnrA box as well as the UC and TnrA regions are indicated in the Pilv-leu region (nt −248/+26). A 7-nt direct repeat, ATAAAAT, was found inside the TnrA box and the most upstream part of the TnrA region, which is likely required for TnrA-mediated negative regulation. (A) Sequential deletion analysis of TnrA-negative regulation. Cells of strains FU676, FU865, FU854, FU866, FU868, FU869, and FU887 carried no deletion, iΔ11, iΔ16, iΔ32, iΔ42, iΔ53, and iΔ63, respectively, in the Pilv-leu region (nt −248/+26), which is fused with lacZ. These strains together with strain FU679 carrying the lacZ fusion (no deletion) and a disrupted tnrA gene were grown to an OD600 of 0.5 under nitrogen-limited conditions, and then β-Gal activity in crude cell extracts was measured. After subtraction of the value for strain FU676 (no deletion at nt −248/+26) from the values for all the internal deletion strains, including strain FU679 (tnrA), the plotted ratios were obtained by subtraction from 1 of the ratio obtained by dividing each subtracted β-Gal value with that of strain FU679 (tnrA). The average rations obtained from at least two independent lacZ-monitoring experiments had at most 10% errors. T-on and T-off denote TnrA-mediated negative regulation (N) was on or off, respectively. (B) Sequential deletion analysis of CA. Cells of lacZ fusion tnrA strains FU679, FU870, FU856, FU871, FU873, FU874, and FU888 carried no deletion, iΔ11, iΔ16, iΔ32, iΔ42, iΔ53, and iΔ63, respectively. These strains together with strain FU724 carrying cre mutations (G-89T C-84T) and tnrA were grown to an OD600 of 0.5 under nitrogen-limited conditions, and then β-Gal activity in crude cell extracts was measured. After subtraction of the β-Gal value for strain FU724 [tnrA cre (G-89T C-84T)] from the values for all the internal deletion strains, including strain FU679 (tnrA), the plotted ratios were obtained by dividing each subtracted value with that for strain FU679. (C) Segmented deletion analysis of the TnrA region downstream of the TnrA box essential for TnrA-negative regulation. Cells of strains FU676, FU1161, FU1160, FU1158, FU1157, FU1162, FU1170, FU1163, and FU1159 carried Δ0 (no deletion), ΔU5, ΔU4, ΔU3, ΔU2, ΔU1, ΔU0, ΔD1, and ΔD2, downstream of the TnrA box, respectively. These strains together with strain FU679 (tnrA) were grown under nitrogen-limited conditions, and then β-Gal activity in crude cell extracts was measured. The plotted ratios were obtained as for panel A. (D) Segmented deletion analysis of TnrA-negative regulation. Cells of strains FU676, FU1172, and FU1173 carried no deletion, ΔC1, and ΔC2, upstream of cre, respectively. These strains together with strain FU679 (tnrA) were grown, and then β-Gal activity in crude cell extracts was measured. The plotted ratios were obtained as described for panel A. (E) Segmented deletion analysis of CA. Cells of tnrA strains FU679, FU1174, and FU1175 carried no deletion, ΔC1, and ΔC2, respectively. These strains together with strain FU724 carrying cre mutations (G-89T C-89T) and tnrA were grown under nitrogen-limited conditions, and then β-Gal activity in crude cell extracts was measured. The plotted ratios were obtained as described for panel B.