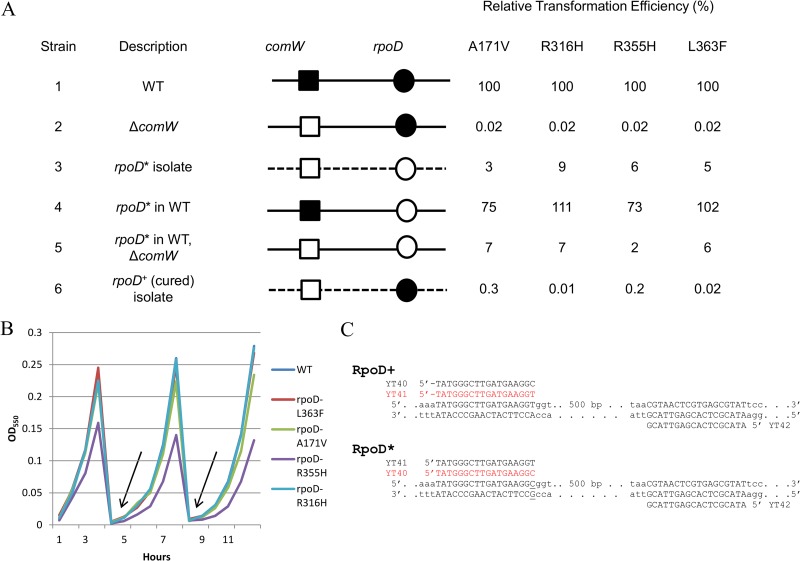
FIG 4.
Linkage between rpoD* mutations and the comW bypass phenotype. (A) Backcross analysis of the bypass phenotypes of four rpoD* isolates. Shown is a comparison of the transformation efficiency of the suppressor isolate (line 3) to those of isolates with rpoD* in a WT background (line 4) and with rpoD* in a transformant ΔcomW WT background (line 5) and a suppressor isolate cured of the rpoD* mutation (line 6). Solid lines, WT genome; dashed lines, genome of isolate recovered from enrichment; ■, comW+; □, ΔcomW; ●, rpoD+; ○, rpoD* (the bypass mutant residues are indicated at the tops of the data columns). For mutations A171V, R316H, R355H, and L363F, the suppressor isolates used were ALT4, ILT1, FLT4, and NYT1, respectively; the strains with backcrossed rpoD* alleles in a ΔcomW WT background were CP2456, CP2458, CP2457, and CP2455; and the rpoD+ cured strains were CP2459, CP2461, CP2460, and CP2462. Standard deviation (SD) values were below 20%. (B) Growth of strains CP2451 to -54 containing rpoD* mutations over 17 generations. The arrows indicate 1:100 dilutions of the exponentially growing culture in fresh medium. (C) Allele-specific alternative primer pairs used for mismatch PCR genotyping of segregants after transformation with 5.5-kb donor rpoD amplicons. The SNP in the A171V mutant is underlined. The productive allele-specific primer for each allele is shown in red.