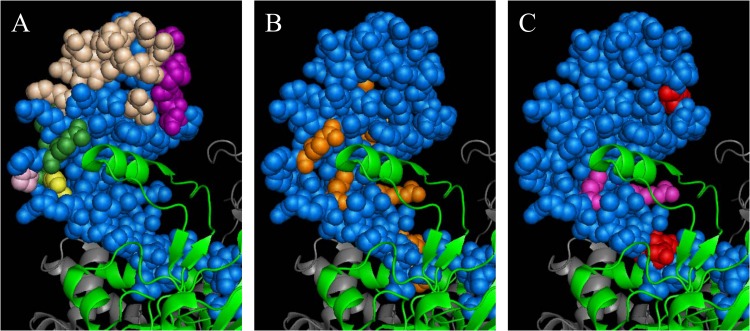
FIG 6.
Locations of DNA-contacting and protein interaction residues, comW bypass residues, and bypass and affinity-affecting residues in region 4 of σA in a holoenzyme. Shown is the crystal structure of a holoenzyme from E. coli (34), Protein Data Bank (PDB) ID 4LJZ. σ70, space-filling blue; β-subunit, green ribbon; β′, gray ribbon. (A) Residues contacting DNA or regulatory proteins in region 4. Beige, DNA-binding residues; dark green, CAP-interacting residues; pink, FNR-interacting residues; yellow, λCI-interacting residues; purple, PhoB-interacting residues (all according to Campbell et al. [26]). (B) comW suppressor residues in region 4. Orange, residues corresponding to comW bypass mutations identified in this study. (C) Bypass and affinity residues from E. coli and T. thermophilus in region 4. Red, residues corresponding to bypass mutations that facilitate the activity of an alternative sigma factor according to Laurie et al. (29); magenta, mutations that reduce σA affinity for RNA according to Dove et al. (27) and Nickels et al. (28).