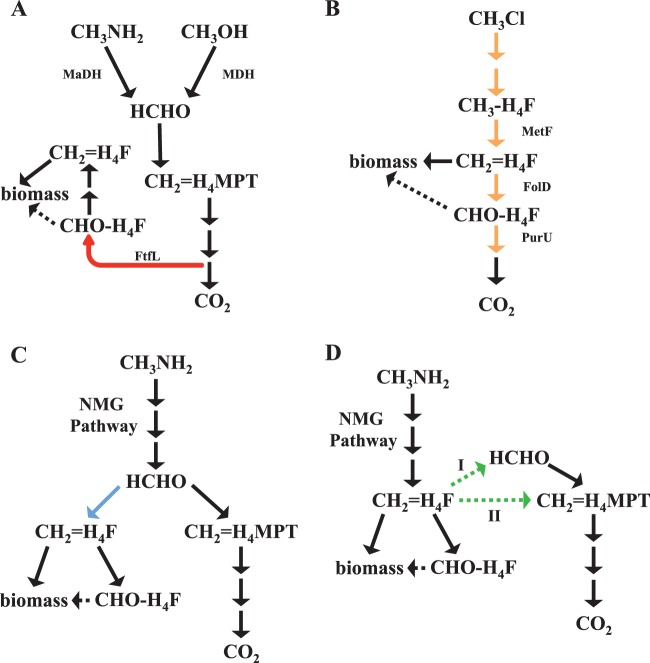
FIG 5.
(A) Schematic representation of methanol dehydrogenase (MDH)- and MaDH-mediated methylamine metabolism in M. extorquens. Primary oxidation generates free formaldehyde, which is oxidized to formate by the H4MPT-dependent C1 dissimilation pathway. Formate is reduced by the H4F-dependent C1 assimilation pathway and assimilated using the serine cycle. The reaction highlighted in red, mediated by FtfL (formyl-tetrahydrofolate ligase), is unique to this topology. (B) Schematic representation of chloromethane metabolism in M. extorquens CM4. Primary oxidation produces H4F derivatives, using an H4F-dependent C1 dissimilation pathway, and completely bypasses formaldehyde production. Methylene tetrahydrofolate (CH2=H4F) is assimilated using the serine cycle. The reactions highlighted in orange, which are catalyzed by enzymes of the H4F-dependent C1 dissimilatory pathway (MetF, FolD, and PurU), are unique to this topology. (C) Schematic representation of the metabolic network involved in methylamine growth if the end product of the N-methylglutamate pathway is formaldehyde. The condensation of formaldehyde and H4F to form CH2=H4F, highlighted in blue, is unique to this topology. (D) Schematic representation of the metabolic network involved in methylamine growth if the end product of the N-methylglutamate pathway is CH2=H4F. CH2=H4F enters the H4MPT-mediated C1 assimilation pathway either by dissociation to formaldehyde (I) or by a direct cofactor switch (II). These two alternate reactions, indicated by green arrows, are unique to this topology.