Abstract
Several members of the obligately aerobic genus Streptomyces are able to reduce nitrate, catalyzed by Nar-type respiratory nitrate reductases. A unique feature of Streptomyces coelicolor A3(2) compared with other streptomycetes is that it synthesizes three nonredundant Nar enzymes. In this study, we show that Nar2 is the main Nar enzyme active in mycelium and could characterize the conditions governing its synthesis. Nar2 was present at low levels in aerobically cultivated mycelium, but synthesis was induced when cultures were grown under oxygen limitation. Growth in the presence of high oxygen concentrations prevented the induction of Nar2 synthesis. Equally, an abrupt shift from aerobiosis to anaerobiosis did not result in the immediate induction of Nar2 synthesis. This suggests that the synthesis of Nar2 is induced during a hypoxic downshift, probably to allow maintenance of a proton gradient during the transition to anaerobiosis. Although no Nar2 could be detected in freshly harvested mature spores, synthesis of the enzyme could be induced after long-term (several days) incubation of these resting spores under anaerobic conditions. Induction of Nar2 synthesis in spores was linked to transcriptional control. Nar2 activity in whole mycelium was strictly dependent on the presence of a putative nitrate transporter, NarK2. The oxygen-dependent inhibition of nitrate reduction by Nar2 was mediated by NarK2-dependent nitrate:nitrite antiport. This antiport mechanism likely prevents the accumulation of toxic nitrite in the cytoplasm. A deletion of the narK2 gene had no effect on Nar1-dependent nitrate reduction in resting spores. Together, our results indicate redox-dependent transcriptional and posttranslational control of nitrate reduction by Nar2.
INTRODUCTION
Streptomyces coelicolor A3(2) is a high-GC-content, Gram-positive, filamentous soil-dwelling bacterium and represents the most important genetic model within the order Streptomycetales (1). These bacteria are reliant on oxygen for growth and undergo a complex life cycle that includes growth as a substrate mycelium, followed by the development of aerial hyphae and the production of spores. While many early studies focused on the regulation of primary metabolism (2), comparatively few studies have addressed the regulation of respiration in Streptomyces (3–5) or what has been referred to as the “anaerobic paradox” (6). According to this paradox, although streptomycetes are unable to grow in the absence of oxygen, their genome nevertheless encodes enzymes whose products are associated with anaerobic metabolism (6). Some of these might contribute to the ability of S. coelicolor to survive extended periods of anoxia (7). The physiological adaption behind this phenomenon is, however, poorly understood. An experimental model that might help shed light on the general mechanisms underlying how Streptomyces species adapt to survive extended periods of oxygen deprivation is based on the respiratory nitrate reductase (Nar) enzymes.
Nar has been shown to play an important role in the establishment of persistence in Mycobacterium tuberculosis, a phylogenetically related actinobacterium that is the causative agent of tuberculosis (8). Respiratory nitrate reduction is important for the survival of M. tuberculosis in macrophages, probably by helping to maintain redox homeostasis during hypoxia (9). While the bacterium cannot grow by nitrate respiration, the ability to respire nitrate helps to maintain the proton gradient during the metabolic remodeling and downshift that the bacterium undergoes to adapt to hypoxia and, eventually, anaerobiosis (8, 10). M. tuberculosis bacteria can survive for many years in this nonreplicating state (11). As a soil bacterium, S. coelicolor potentially has to deal with similar hypoxic or anoxic conditions, especially during extended periods of wet conditions, and it is conceivable that it enters a metabolic state analogous to that in M. tuberculosis (7). Due to the fact that in M. tuberculosis nitrate reduction contributes significantly to hypoxic survival, it is possible that S. coelicolor's contingent of Nar enzymes has a similarly important function.
In contrast to M. tuberculosis, the genome of S. coelicolor A3(2) encodes three nonredundant, active Nar enzymes (12). Nar1 has recently been characterized as the first known spore-specific Nar enzyme (13) and is always present in a ready-to-use mode in mature spores, but it is not found in mycelium. The in vivo activity of Nar1-dependent nitrate reduction is initiated only in the absence of ambient oxygen and results in the stoichiometric release of nitrite when exogenous nitrate is available (12, 13). Less is known about the Nar2 and Nar3 enzymes, other than the fact that they are primarily active in mycelium and not in spores (12). It is conceivable that, like for Nar1, the regulation of the synthesis of Nar2 and Nar3 differs from the regulatory mechanisms controlling Nar enzyme synthesis in other microorganisms. Therefore, the current study focuses on a detailed characterization of the Nar2 enzyme of S. coelicolor A3(2). We demonstrate that nitrate reduction by Nar2 occurs only in the absence of oxygen and that this requires a NarK-type transporter which is functional only in mycelium. Moreover, Nar2 synthesis is induced during oxygen limitation rather than after an abrupt shift to anaerobiosis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The media and culture conditions for Escherichia coli and S. coelicolor were the same as those described previously (14, 15). The strains used in this study are listed in Table 1. E. coli DH5α (Stratagene) was used as a host for cosmids and for plasmid constructions. E. coli ET12567(pUZ8002) (16) and JTU007(pUZ8002) (17) were the nonmethylating plasmid donor strains used for intergeneric conjugation with S. coelicolor strain M145 (20). Apramycin (Apra; 25 μg ml−1), carbenicillin (Carb; 100 μg ml−1), chloramphenicol (CLM; 25 μg ml−1), kanamycin (Kan; 25 μg ml−1), spectinomycin (Spc; 25 μg ml−1), or hygromycin (Hyg; 25 μg ml−1), all from Sigma, was added to the growth medium when required.
TABLE 1.
Strains and vectors used in this study
| Strain, plasmid, or cosmid | Genotype and characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Streptomyces coelicolor A3(2) | ||
| M145 (wild type) | SCP1− SCP2− | 15 |
| NM3 (Δnar-2) | M145 ΔSCO0216-ΔSCO0219::aac(3)IV (deletion of 6,531 bp removing narG2H2J2I2) | 12 |
| NM24 (Δnar-1) | M145 ΔSCO6535-ΔSCO6532::aadA (deletion of 6,209 bp removing narG1H1J1I1) | 13 |
| NM27 (Δnar-3) | M145 ΔSCO4947-ΔSCO4950::aadA (deletion of 6,497 bp removing narG3H3J3I3) | 13 |
| NM29 (Δnar-1 Δnar-2) | NM4 ΔSCO6535-ΔSCO6532::aadA (deletion of 6,209 bp removing narG3H3J3I3) | 13 |
| NM59 (Δnar-1 Δnar-3) | NM39 ΔSCO6535-ΔSCO6532::aadA (deletion of 6,209 bp removing narG3H3J3I3) | 13 |
| NM68 (Δnar-2 Δnar-3) | NM3 ΔSCO4947-ΔSCO4950::aadA (deletion of 6,497 bp removing narG3H3J3I3) | 12 |
| NM92 (Δnar-1 Δnar-2 Δnar-3) | NM90 ΔSCO6532-ΔSCO6535::aadA (deletion of 6,209 bp removing narG1H1J1I1) | 12 |
| COE285 | M145 SCO0213::Tn5062 (ΔnarK2) | This study |
| COE404 | COE285 with integrative vector pMSnarK2 (SCO0213+) | This study |
| COE301 | M145 SCO2959::Tn5062 (ΔnarK1) | This study |
| Escherichia coli | ||
| DH5α | F− ϕ80dlacZΔM15 endA recA hsdR(rK− mK−) supE thi-1 gyrA relA Δ(lacZYA-argF)U169 | Laboratory stock |
| ET12567(pUZ8002) | Δdam dcm with trans-mobilizing plasmid pUZ8002 | 16 |
| JTU007(pUZ8002) | Δdam Δdcm with trans-mobilizing plasmid pUZ8002 | 17 |
| Rosetta(DE3)/pLysS | F− ompT hsdSB(rB− mB−) gal dcm λ(DE3 [lacI lacUV5-T7 gene 1 ind1 Sam7 nin5])/pLysSRARE (CLMr) | Novagen |
| Plasmids and cosmids | ||
| SCJ12.2.03_04081112EO | Cosmid SCJ12 disrupted in SCO0203 with Tn5062 | 18 |
| SCJ12.1.F10_040807014A | Cosmid SCJ12 disrupted in SCO0213 with Tn5062 | 18 |
| pMS82 | ΦBT1 attP-int-derived integration vector for conjugal transfer of DNA from E. coli to Streptomyces (Hygr) | 19 |
| pMSnarK2 | pMS82 SCO0213 (with 236-bp upstream and 227-bp downstream sequences) | This study |
| pET30a | (+) Expression plasmid, Kanr | Novagen |
| pET30-narG2 | Like pET30a(+) but narG2 positive | This study |
S. coelicolor A3(2) wild-type strain M145 and mutant derivatives (Table 1) were grown on R2YE, SFM, R5, LB, or Difco nutrient broth (DNB) agar medium, as indicated below, or in liquid tryptic soy broth (Oxoid) or DNB supplemented with antibiotics to maintain selection when appropriate. The growth medium compositions and the standard culture techniques used have been described previously (15).
Construction of gene disruption mutants.
A cosmid (Table 1) with a transposon insertion (Tn5062) in the SCO0213 (narK2) gene was introduced into E. coli ET12567(pUZ8002) or JTU007(pUZ8002) by electroporation and then transferred to Streptomyces by conjugation (15). Exconjugants with double crossovers were selected for Kans and Aprar. S. coelicolor mutant strains were confirmed by PCRs and by Southern hybridization (data not shown).
Complementation of gene disruptions.
A 1,239-bp DNA fragment corresponding to the coding region of SCO0213 (narK2) plus 236 bp of the upstream flanking sequence and 227 bp of the downstream flanking sequence, including potential regulatory elements, was amplified by PCR using oligonucleotides 0213F (5′-CATGGGGTCCCGGCTTCCAG-3′) and 0213R (5′-GGGCCATCGCGGGAAAGAGC-3′) and cloned between the HindIII and KpnI restriction sites of integrative Streptomyces vector pMS82 (19) (Table 1) to deliver pMSnarK2. Plasmid pMSnarK2 was introduced into M145 via conjugation using the plasmid-containing E. coli strain ET12567(pUZ8002), which mobilizes the oriT-containing plasmid pMS82 for conjugation in trans (Table 1).
Culture conditions adapted for physiological studies.
S. coelicolor A3(2) strains were grown as highly disperse liquid cultures in Duran-F tubes with MOPS (morpholinepropanesulfonic acid)-buffered Trypticase soy broth (TSB) as described before (21). For cultivations in which oxygen concentrations were monitored, the medium was flushed continuously with compressed air through a needle (0.8 mm, 20 to 40 liters/h, for Duran-F tubes) or sintered glass (60 liters/h, for Erlenmeyer flasks). Hypoxic cultures were individually tested for oxygen concentration by empirically determining the relationship between the cell amount, the surface-to-volume ratio (of the vessel and medium), as well as the stirring or shaking parameters. For defined oxygen-limited cultivations, the medium was flushed with a defined air-gas mixture, which was achieved using a gas mixer (G21A6-BA0; Aalborg Instruments, Orangeburg, NY, USA), with the air-gas mixture consisting of between ca. 10% air plus 90% nitrogen or ca. 30% air plus 70% nitrogen. Anoxic conditions were achieved by flushing the vessels with nitrogen for 5 to 10 min. In order to grow cell aggregates (minipellets) of defined size, highly dispersed exponential precultures were transferred to fresh medium at a ratio of 4,000 cell amount equivalents (CAE) (21) per 50 ml of TSB. Afterwards, they were incubated in Erlenmeyer flasks or 24-well plates with orbital shaking (170 rpm) until the desired size (0.25 to 0.5 mm) of aggregate was attained.
For the in vivo assay in mycelium, small amounts of cells (determined in parallel) were transferred to anoxic Hungate vials with fresh TSB medium supplemented with 10 mM nitrate, and these were incubated for at least 30 min.
Standardized 15-h-old exponential-phase cultures (20 ml) were inoculated with 2 ml of a standard mycelium suspension. This suspension was prepared from a highly disperse preculture by the determination of the cell pellet size after centrifugation (2,000 × g, 10 min, 6°C). A pellet volume of 200 μl was diluted to 10 ml. Afterwards, the cultures were incubated in Duran-F vials for 15 h, as described before (21).
Standard liquid cultures were incubated in small reaction tubes (volume, 2 ml) or 24-well plates (21). Therefore, 0.5 ml of double-concentrated incubation buffer (usually 25 mM MOPS-NaOH, pH 7.2, 10 mM NaNO3) was supplemented with washed mycelium and water up to a 1-ml volume.
Oxygen monitoring during the growth and incubation assays was done using oxygen-dependent luminescence sensor spots (PyroScience, Aachen, Germany). These spots were affixed to the inner side (glass wall) of vials and were noninvasively connected to an optical oxygen meter (FireStingO2). Signals were analyzed using FireSting Logger software (PyroScience, Aachen, Germany).
Methylene blue adsorption (MBA) measurements for calculating CAE, as well as small-scale growth curves, were performed as described before (21).
Analysis of nitrite production in spore suspensions.
On solid medium, spore suspensions (5 μl of a suspension with an optical density at 450 nm [OD450] of 20) were dropped onto LB agar (3 ml) with or without 200 μg ml−1 chloramphenicol in a 24-well plate and incubated immediately under anaerobic conditions for 3 days at 30°C. When required, sodium nitrate (5 mM) was included in the medium. In liquid medium, spore suspensions (20 ml of a suspension with an OD450 of 5) were anaerobically incubated with 50 mM MOPS-NaOH buffer and 5 mM nitrate in serum flasks. Excreted nitrite was determined colorimetrically as described below, and the presence of nitrite was revealed as a dark violet staining of the well (12).
Cloning of narG2 and overproduction and purification of the His-tagged NarG2 polypeptide.
The construction of the narG2 expression plasmid pET30-narG2 was achieved by amplifying the complete narG2 gene using genomic DNA from S. coelicolor M145 as the template. Amplification was performed using the oligonucleotides NarG2/NdeI_forward (5′-GGT GGT CAT ATG GAG AAC GAT CAG AAC GCA CGC-3′) and NarG2/XhoI_reverse (5′-GGT GGT CTC GAG GTA CTC CAC TCG CTG GTC GCG-3′), which introduced XhoI and NdeI restriction sites. The PCR was carried out using Herculase II Fusion DNA polymerase (Agilent Technologies). The resulting 3,720-bp DNA fragment was cloned into an XhoI- and NdeI-digested pET30a(+) vector (Novagen) to deliver pET30-narG2.
For overproduction of His-tagged NarG2, cultures of E. coli Rosetta(DE3)/pLysS (Novagen) were used. The overproduction and purification were performed as described for narG1 (13). The purified His-tagged NarG2 was stored at −20°C.
Antibody preparation and Western blotting.
Antibodies against a 14-amino-acid peptide (NSPRHYGDERLHED, amino acid positions 1066 to 1079) in the NarG2 polypeptide were prepared commercially (Seqlab, Göttingen, Germany). To minimize unspecific cross-reactions, antiserum raised against the NarG2 peptide was treated by depleting unspecific cross-reacting antibodies using a crude extract derived from mycelium of the nar-2-knockout mutant NM3. The procedure for the depletion was carried out as described before (13). The supernatant obtained from depletion was used as the primary antibody for Western blot analysis. The treated antiserum was used in the dilution range of 1:25 to 1:150.
PAGE and immunoblotting.
Aliquots (60 μg of protein) from the indicated subcellular fractions were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using 7.5% (wt/vol) polyacrylamide gels (22) and transferred to nitrocellulose membranes as described previously (23). Purified anti-NarG2 antibodies (dilution, 1:25) were used to detect NarG2 polypeptide in the crude extracts of spores. Secondary antibody conjugated to horseradish peroxidase was obtained from Bio-Rad. Visualization was done by use of the enhanced chemiluminescent reaction (Stratagene).
RNA extraction from exponential- and stationary-phase mycelium.
RNA extraction from mycelium and purification were performed as described previously (13). Reverse transcription-PCR (RT-PCR) and cDNA preparation were also performed as described previously (13). The oligonucleotides used to monitor narG2 transcript levels were RT-narG2_212fw (CGTGCTCCTGGATGGTGTACG) and RT-narG2_821rv (GTGACGGGGATGTTGGAGCCC), and those used to detect 16S rRNA gene transcripts, which was done to check the quality of the RNA used for cDNA synthesis, were the same as those described in reference 13.
Other methods.
Quantitative determination of nitrite in culture supernatants and mycelial dry weight measurement were performed exactly as described previously (12). Nitrate reductase enzyme activity in crude extracts was determined using the continuous assay procedure with reduced dithionite and benzyl viologen (0.4 mM) at 30°C, as described previously (12, 24). Crude cell extracts of S. coelicolor mycelium were prepared by sonication of mycelium in buffer (100 mM potassium phosphate, pH 7.2). Resuspended mycelium (2 to 5 ml) was sonicated four times for 3 min each time at 30 W (pulses of 0.5 s on and 0.5 s off) using a Sonoplus sonifier with a Sonotrode KE76 tip (Bandelin, Berlin, Germany).
Standard cloning methods (25) were used.
RESULTS
Nar2 is the main respiratory nitrate reductase in mycelium.
In order to determine which Nar enzyme is responsible for nitrate reduction in mycelium, Nar enzyme activity was analyzed in crude extracts derived from mycelium in exponentially growing cultures of different nar operon mutants (Table 2). Results presented previously (13) demonstrated that Nar1 is inactive in mycelium. Crude extracts of strains NM3 (Δnar-2) and NM29 (Δnar-1 Δnar-2) had activities that were more than 95% lower than the activity in the extract of wild-type strain M145 (Table 2), while an extract derived from NM59 (Δnar-1 Δnar-3) retained approximately 70% of the activity measured in M145 extracts. No Nar enzyme activity was detected in an extract derived from strain NM92 (Δnar-1 Δnar-2 Δnar-3) (Table 2), which lacks all three Nar enzymes (12). These results indicate that Nar2 is mainly responsible for the nitrate-reducing activity in mycelial cultures.
TABLE 2.
Nitrate reductase activity in extracts of mycelium
| Strain of S. coelicolor used for extract preparation | Nitrate reductase sp act (mU)a |
|---|---|
| M145 (wild type) | 17.26 ± 3.13 |
| NM3 (Δnar-2) | 0.75 |
| NM29 (Δnar-1 Δnar-2) | <0.1 |
| NM59 (Δnar-1 Δnar-3) | 12.38 |
| NM92 (Δnar-1 Δnar-2 Δnar-3) | <0.01 |
| COE285 (ΔnarK2) | 14.27 |
| COE404 (ΔnarK2 + narK2) | 14.10 |
Experiments were performed a minimum of two times, and each time, extracts were measured two or three times. Data represent the average of the mean and are given as nmol nitrate reduced min−1 mg protein−1.
Aliquots of these crude extracts were loaded on a nondenaturing polyacrylamide gel, and after electrophoresis the gel was stained to reveal Nar enzyme activity (Fig. 1A). A single activity-staining band was detected only in extracts from strains whose genomes encoded a Nar2 enzyme; no activity band was identified in extracts from strain NM3 (Δnar-2), NM29 (Δnar-1 Δnar-2), NM68 (Δnar-2 Δnar-3), or NM92 (Δnar-1 Δnar-2 Δnar-3). Western blot analysis after SDS-PAGE of the same extracts using antibodies directed against a NarG2-specific peptide confirmed that NarG2 could be detected only in extracts of strains M145, NM24 (Δnar-1), NM27 (Δnar-3), and NM59 (Δnar-1 Δnar-3) (Fig. 1B).
FIG 1.
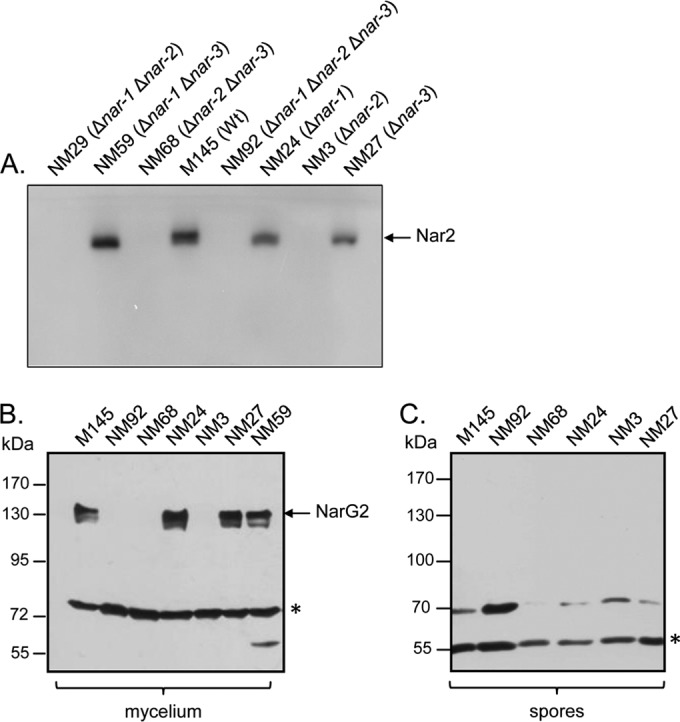
Nar2 is the main nitrate reductase active in growing mycelium. (A) In-gel Nar activity was analyzed by separating aliquots (60 μg) of detergent-treated crude extracts in a nondenaturing polyacrylamide gel (10% [wt/vol]) and subsequently staining with a buffer containing dithionite-methyl viologen (MV) and nitrate (see Materials and Methods). Nar2 activity is indicated, and the inverse of the zymogram is shown. Wt, wild type. (B) Western blot analysis of NarG2 in crude extracts (60 μg) from exponentially grown mycelium was analyzed by separating the polypeptides by SDS-PAGE (7.5% [wt/vol] polyacrylamide) using peptide antibodies specific for the NarG2 catalytic subunit. NarG2 migrates at approximately 139 kDa. An unidentified cross-reacting polypeptide (marked with an asterisk) migrating at approximately 70 kDa acted as a loading control. (C) A Western blot of crude extracts (60 μg) derived from spores of the indicated S. coelicolor nar deletion mutants is shown. Polypeptides were separated by SDS-PAGE (7.5% [wt/vol] polyacrylamide) and analyzed with NarG2-specific peptide antibody exactly as described for the mycelium extracts. An unidentified cross-reacting band (marked with an asterisk) migrating at 55 kDa acted as the loading control.
Oxygen limitation induces the synthesis of Nar2-dependent nitrate respiration in mycelium.
In a previous study (12), nitrate reduction was observed in both aerobically grown streptomycete cultures and anaerobically incubated mycelium. It was suggested that Nar2 was mainly responsible for the aerobic activity, even though for all intents and purposes it is assumed to be an anaerobic process. Due to the tendency of S. coelicolor to grow as aggregates, especially in the presence of large amounts of mycelium, these aggregates are likely to become oxygen limited, possibly explaining the Nar activity observed previously (12). Therefore, it was decided to standardize mycelial growth with respect to the oxygen concentration for the analysis of Nar2 activity (Fig. 2A). If mycelium was grown with more than 2 mg liter−1 O2 (air flushed), no significant nitrate reduction was detectable in whole mycelium. However, if the same mycelium was grown for a further 2 h at a lower oxygen concentration (hypoxic conditions), the mycelium released 4 to 8 nmol nitrite h−1 CAE−1 into the medium (Fig. 2A). A crude extract derived from this mycelium had Nar activity of 14.8 mU mg−1. These nitrate-reducing activities in whole mycelium of hypoxic cultures were similar to those observed previously (12) when cultures were grown by standard techniques (i.e., in shaking flasks and tubes) without the use of an air-sparging system and with what was referred to as exponentially growing mycelium. In that study (12) it was suggested that Nar2 is the main Nar enzyme active during this growth stage.
FIG 2.
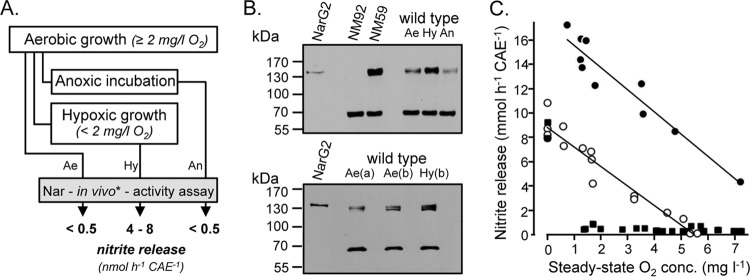
Nitrate reductase activity in substrate mycelium of S. coelicolor A3(2) M145 (wild type). (A) A scheme for preparation of cell material used in the experiments whose results are shown in panels B and C. The in vivo nitrate-reducing activities for the respective samples are shown. Ae, aerobic growth; Hy, hypoxic incubation; An, anaerobic incubation. The asterisk denotes measurement of nitrite released. (B) Detection of NarG2 with peptide antibodies raised against the catalytic subunit NarG2. (Top) Crude extracts (50 μg) derived from the cultures described in panel A; (bottom) crude extracts (50 μg) derived from cultures grown with defined and stable oxygen concentrations of ca. 2.2 mg O2 liter−1 (aerobic growth) and ca. 0.7 mg O2 liter−1 (hypoxic incubation). (a), the mycelia were fully dispersed; (b) the mycelia were present as micropellets. (C) Nitrite release by differentially prepared mycelium (see Materials and Methods for details) incubated in TSB in the presence of 200 μg/ml chloramphenicol. Nitrate was added after reaching steady state. Each point represents an individual measurement with dispersed mycelium (squares), 0.25-mm mycelial pellets (open circles), and 0.5-mm mycelial pellets (filled circles). Anoxic conditions were achieved by closing the vials with gastight stoppers and flushing with nitrogen.
In contrast to the increase in nitrate-reducing activity detected in the mycelium of cultures shifted to hypoxic conditions (Fig. 2A), surprisingly, upon an abrupt shift of the cultures grown with oxygen sparging for 2 h to anoxic conditions, no Nar2-dependent nitrate-reducing activity could be detected (Fig. 2A). Western blot analysis of crude extracts derived from these mycelia revealed that while low-level NarG2 synthesis was detectable both in aerobically cultivated mycelium and in mycelium that had been abruptly shifted to anoxic conditions, NarG2 levels in crude extract from the mycelium of the hypoxic cultures (<2 mg dissolved O2 liter−1) was significantly increased (Fig. 2B, top). These data suggest a two-step induction of Nar2 synthesis: in addition to the low-level Nar2 synthesis evident in aerobically cultivated mycelium, a further hypoxic induction of Nar2 synthesis concomitant with Nar2-dependent nitrate respiration in mycelium occurs. To confirm this finding, we grew S. coelicolor wild-type strain M145 under defined and constant oxygen-limited conditions (achieved by quantitative gas mixing; see Materials and Methods) employing two oxygen concentrations of 0.7 mg/liter (2%) and 2.2 mg/liter (6%) dissolved O2. Conditions were also employed to maintain the growth of the bacterium as highly disperse mycelium or micropellets. A decrease in the oxygen concentration to 0.7 mg/liter dissolved O2 (hypoxic conditions) resulted in significantly increased Nar2 synthesis (Fig. 2B, bottom).
In addition to the oxygen-dependent Nar2 synthesis, our preliminary results suggested an oxygen-dependent activation of respiratory nitrate reduction even if the Nar2 enzyme was present (Fig. 2B). This is important with regard to the findings that nitrate reduction occurs only under local hypoxic or anoxic conditions, for example, in highly aggregated mycelial cultures. To demonstrate that this was indeed the case, we analyzed chloramphenicol-treated mycelium (grown hypoxically) for its ability to reduce nitrate in the presence of different oxygen concentrations in the medium; it has been demonstrated that oxygen inhibits nitrate reduction (see above) (13). While highly disperse mycelium reduced nitrate to nitrite only in the complete absence of oxygen, mycelium aggregated to form minipellets (0.25 or 0.5 mm in diameter) reduced nitrate at a rate inversely correlated with the oxygen level in the medium (Fig. 2C). This result suggests the presence of an oxygen gradient within the pelleted aggregate and, hence, that this causes anaerobic nitrate reduction during aerobic cultivation of S. coelicolor A3(2).
Nar2-dependent nitrate respiration by intact mycelium requires the predicted nitrate:nitrite antiporter NarK2 and the absence of oxygen.
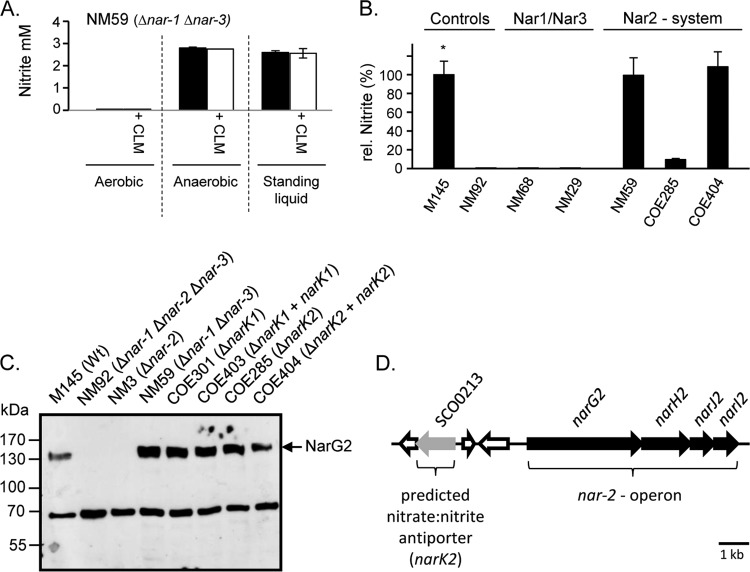
We could show previously that anaerobic incubation was required to observe the maximal reduction of nitrate by mycelial cultures of strain M145 (12). In order to standardize the conditions for measuring nitrate reduction, we used a 3-day staged cultivation procedure culminating in a 15-h growth period in TSB medium to achieve reproducible cell densities and aggregation sizes of mycelium (see Materials and Methods for details) (21). Aliquots of these mycelia (ca. 250 CAE) from strain NM59 (only Nar2 was present) were used for determination in vivo of Nar2-dependent nitrate reduction in MOPS buffer (pH 7.0) with nitrate (10 mM) but without other substrates (Fig. 3A). Aerobic incubation failed to result in any measureable nitrite production. In contrast, mycelium incubated under an N2 atmosphere reduced ca. 3 mM nitrate within 2.5 h (Fig. 3A). Addition of chloramphenicol had no effect upon the levels of nitrate reduction, indicating that de novo protein synthesis was not required. A similar level of nitrate reduction was also noted when the mycelium was incubated in a standing liquid assay, whereby it was not necessary to place it under an N2 atmosphere (Fig. 3A).
FIG 3.
Oxygen-inhibited and NarK2-dependent reduction of nitrate by the mycelium of S. coelicolor. (A) Nitrite production by the mycelium of S. coelicolor strain NM59 (Δnar-1 Δnar-3) after 2.5 h of incubation in MOPS buffer in the presence of 10 mM nitrate was monitored. Mycelium was incubated aerobically, anaerobically, or in standing liquid culture. One of each variant was additionally incubated with 200 μg/ml chloramphenicol (CLM). (B) Relative (rel.) nitrite release of mutant strains of S. coelicolor A3(2) in comparison to that of wild-type strain M145 (*, 100% relative nitrite release is equivalent to approximately 2 mM nitrite). Nitrite was determined after 2.5 h of incubation in MOPS buffer with 10 mM NO3−. The incubation was carried out with standing liquid cultures. (C) Western blot analysis with anti-NarG2 peptide antibodies of extracts (60 μg) of mycelium derived from the indicated mutants. After separation in SDS-polyacrylamide gels (7.5% [wt/vol] polyacrylamide), the samples were probed with polyclonal peptide antibodies specific for NarG2. The migration position of NarG2 (139 kDa) is shown on the right. An unidentified cross-reacting band migrating at approximately 70 kDa acted as a loading control. (D) Schematic representation of the genetic locus upstream of the narG2H2J2I2 operon (SCO0216 to SCO0219) in the S. coelicolor A3(2) genome.
Using the standing liquid culture approach, the nitrate reduction assay was performed with mycelium of wild-type strain M145, and the activity determined was compared with the activities measured in various nar-knockout mutants (Fig. 3B). Both M145 and NM59 (Δnar-1 Δnar-3) exhibited similar activities, indicating that nitrate reduction under these conditions was due to Nar2. As controls, no nitrate reduction was observed in the mycelium of strain NM92 (Δnar-1 Δnar-2 Δnar-3), NM29 (Δnar-1 Δnar-2), or NM68 (Δnar-2 Δnar-3).
The fact that nitrate reduction in whole mycelium was inhibited by oxygen, yet the Nar2 enzyme was present, suggested either that Nar2 enzyme activity was the target of oxygen or that nitrate transport was oxygen sensitive. Oxygen-regulated nitrate transport has already been observed in spores (13), and Nar2 activity in crude extracts is unaffected by aerobic incubation (see below); therefore, it is plausible that the delivery of nitrate across the cytoplasmic membrane of the mycelium might be inhibited in the presence of oxygen. Examination of the narG2H2J2I2 locus on the S. coelicolor chromosome (Fig. 3D) revealed the presence of a gene (SCO0213) whose product has approximately 37% amino acid identity to a putative NarK nitrite extrusion protein from Bacillus subtilis (26) (EMBL accession number z49884) and 22% identity with the NarK nitrate:nitrite antiporter from Escherichia coli (27). An S. coelicolor strain, COE285 (ΔSCO0213 ΔnarK2), carrying a disruption in the SCO0213 gene was constructed (see Materials and Methods), and mycelium was prepared as described above and analyzed using the standing liquid assay for nitrate reduction (Fig. 3B). The results revealed that mycelium from strain COE285 reduced only approximately 10% of the nitrate compared with the amount reduced by mycelium from M145. Reintroduction of the SCO0213 gene onto the chromosome via the integrative plasmid pMS82 (19) restored the ability of intact mycelium of the strain to reduce nitrate to nitrite. Henceforth, gene SCO0213 is referred to as narK2.
Crude extracts derived from strains COE285 (ΔnarK2) and COE404 (Table 1) were assayed for Nar2 enzyme activity, and extracts of both strains exhibited activities in the range of 80% of the activity of wild-type strain M145 (Table 2). The same Nar2 enzyme activity was measured regardless of whether the extracts had been incubated aerobically or anaerobically. Analysis of these extracts by Western blotting confirmed that the NarG2 polypeptide was present at levels equivalent to those in extracts of M145 (Fig. 3C). Extracts derived from a further strain (COE301) carrying a disruption in the SCO2959 gene, which encodes a putative membrane protein exhibiting 39% amino acid sequence identity to NarK from E. coli and which we term NarK1, also revealed levels of NarG2 polypeptide similar to those of the wild type (Fig. 3C). Mycelium derived from strain COE301 had nitrate reduction activity similar to that of M145 (data not shown). Therefore, the putative NarK1 gene product is not involved in Nar2-dependent nitrate reduction in mycelium.
Finally, the ability of spores from strain COE285 (ΔnarK2) to reduce nitrate to nitrite anaerobically (13) was tested. Nitrate reduction by spores from strain COE285 (0.42 nmol NO2− min−1 ml−1 OD unit−1) was similar to that by spores from the wild type (0.43 nmol NO2− min−1 ml−1 OD unit−1) (13), and therefore, NarK2 is specifically involved in nitrate reduction in mycelium.
Long-term anaerobic incubation of resting spores induces Nar2-dependent nitrate respiration.
In contrast to the findings for Nar1, previous studies demonstrated that neither Nar2 nor Nar3 is active in spores during a 5-h anaerobic incubation period (13). Western blot analysis of extracts derived from aerobic, freshly harvested resting spores of strains M145, NM92 (Δnar-1 Δnar-2 Δnar-3), NM68 (Δnar-1 Δnar-2), NM24 (Δnar-1), NM3 (Δnar-3), and NM27 (Δnar-3) revealed that no NarG2 polypeptide could be detected (Fig. 1C). Nevertheless, RT-PCR analysis of total RNA derived from freshly harvested spores revealed the presence of narG2 transcripts (Fig. 4). In order to clarify whether these transcripts could function as the templates for rapid synthesis of Nar2 upon germination, we incubated anaerobically arrested spores for up to 3 days on a solid medium with nitrate (Fig. 5A). After this incubation, Nar activity was determined qualitatively by visualization of nitrite production using a nitrite detection assay. A strain without nar genes (NM92) and anaerobically incubated spores without nitrate in the medium served as negative controls. A further incubation included the protein synthesis inhibitor chloramphenicol (Fig. 5A, left). As shown previously (13), spores of the wild type and strain NM68 (Δnar-2 Δnar-3) reduced nitrate to nitrite (Fig. 5A). This was due to Nar1, which is constitutively present in spores (13). Unexpectedly, however, nitrite levels were higher in spores of the wild type (M145) and in a strain lacking nar-1 genes (NM24) than in NM68 (Δnar-2 Δnar-3) (Fig. 5A, right). Moreover, this Nar1-independent nitrate reduction required de novo protein synthesis because it was prevented by addition of chloramphenicol (Fig. 5A, left). It was apparent from the results for strains lacking Nar3 (data not shown) that this activity is associated with Nar2, and so strain NM59 (Δnar-1 Δnar-3) was used for further quantitative analyses (Fig. 5B).
FIG 4.
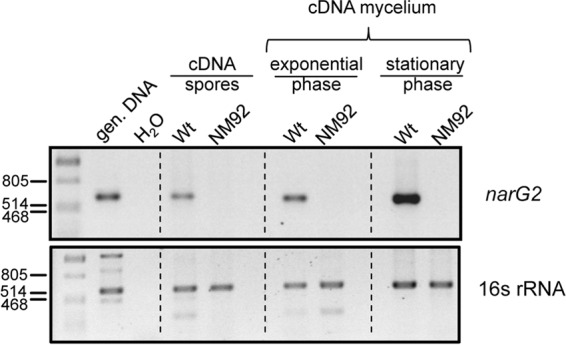
Detection of mRNA transcripts of narG2 in resting spores, exponential-phase mycelium, and late-exponential-phase mycelium of S. coelicolor. Total RNA was isolated from the spores and mycelium of strains M145 (wild type) and NM92 (Δnar-1 Δnar-2 Δnar-3) and was used for cDNA synthesis, as described in Materials and Methods. Aliquots (100 ng) of cDNA were then used in PCRs with the appropriate oligonucleotide combinations (see Materials and Methods). Genomic DNA (gen. DNA) from M145 acted as a positive control, and H2O acted as a negative control, in which water replaced the cDNA library in the PCR. Numbers on the left are molecular sizes (in base pairs).
FIG 5.
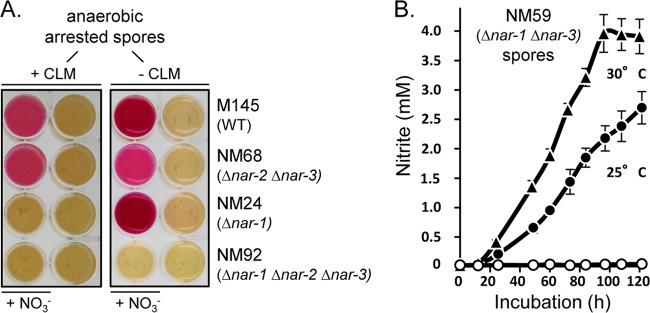
Qualitative (A) and quantitative (B) analysis of Nar2-dependent nitrite production by developmentally arrested (nongerminated) spores of S. coelicolor. (A) Nar-dependent nitrate reduction was determined for the S. coelicolor strains, as indicated in Materials and Methods. Spore suspensions (5 μl of a suspension with an OD450 of 20) were dropped on the plates, which were immediately incubated under anaerobic conditions for 3 days at 30°C. The plate on the right was prepared and processed in the same manner as the plate on the left but without chloramphenicol. Where indicated, 200 μg ml−1 chloramphenicol and 5 mM sodium nitrate (NO3−) were included in the medium. The presence of nitrite is revealed as a dark violet staining of the well. (B) Nar2-dependent nitrate reduction by NM59 (Δnar-1 Δnar-3) was determined quantitatively in anaerobic spore suspensions (suspensions with OD450s equivalent to 5) at 25°C (filled circles) and at 30°C (filled triangles). Spore suspensions were incubated in MOPS-NaOH with 5 mM nitrate for 6 days under a nitrogen atmosphere. A control spore suspension containing 200 μg ml−1 chloramphenicol (open circles) was incubated anaerobically, and a further spore suspension was incubated aerobically (open triangles, which are hidden by the open circles).
Spore suspensions (OD450 = 2.5) were incubated under N2 in MOPS buffer with 5 mM nitrate for 5 days. No germination occurs under these conditions (12, 13). While no release of nitrite was observed in a control incubated aerobically for 5 days, the first significant amounts of nitrite were produced and detected after 1 day in the absence of oxygen (Fig. 5B). Thereafter, a linear release of nitrite at 0.13 nmol NO2− h−1 min−1 OD unit−1 (30°C) was measured. This stable nitrate reductase activity of Nar2 was approximately 70% lower than the Nar1-dependent in vivo activity measured in spores during the first 5 h of an anaerobic incubation of NM68 (Δnar-2 Δnar-3) and the wild type (13). Notably, an anoxic preincubation for 48 h without nitrate did not affect the rate of nitrate reduction (data not shown), indicating that nitrate is apparently not required for the induction of enzyme synthesis. The reducing equivalents for this nitrate respiration in spores were provided solely from internal storage compounds. Moreover, addition of glucose, mannose, or trehalose or the incubation of the spores in rich medium did not result in higher Nar activity (data not shown). Nevertheless, in contrast to the findings for Nar1, the results for controls incubated in the presence of chloramphenicol revealed that de novo protein synthesis was required for the synthesis of Nar2 in spores (Fig. 5B). Another control for anaerobic spore incubations was carried out with rifampin to prevent transcription. Although Nar2 transcripts were detectable in freshly harvested spores (Fig. 4), no nitrate reduction in anaerobically incubated spores was detectable. This indicates that transcription of the nar-2 operon or the expression of another component is necessary for the establishment of Nar2-dependent nitrate respiration in intact spores.
Nitrite production by mycelium causes self-inflicted toxicity.
While anaerobically incubated spores did not germinate even after 5 days, aerobically incubated spores typically germinate in the presence of substrates within 5 to 15 h. It was noted previously that such germlings show in vivo nitrate reduction activity when incubated anaerobically (12). We then analyzed Nar-dependent nitrite release in germlings (pregerminated spores) grown on LB agar using defined nar-knockout mutants (Fig. 6A, left) to determine which Nar enzyme was responsible for the activity. The results shown in Fig. 6A clearly demonstrate that this nitrate reduction activity in germlings was due to Nar2.
FIG 6.
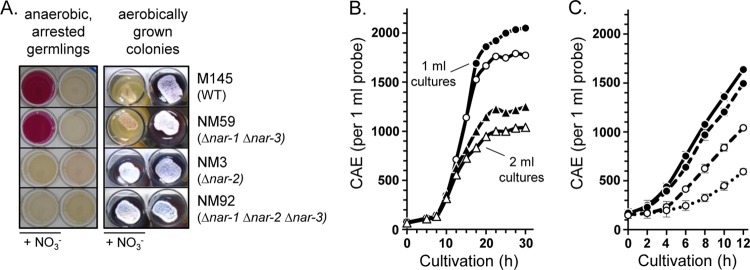
Nitrite release by vegetative stages of S. coelicolor A3(2) and its influence on growth of substrate mycelium on solid medium (A) as well as in liquid culture (B, C). (A) Nar-dependent nitrate reduction in germlings of the strains indicated on the right. For the determination of nitrite production (the two columns on the left), spore suspensions (5 μl of a suspension with an OD450 of 20) were incubated aerobically for 10 h. After germination, the plates were incubated under anaerobic conditions for 3 days at 30°C. In the wells labeled NO3−, 5 mM sodium nitrate was included in the medium. Colony development (the two columns on the right) was analyzed by placing spore suspensions (5 μl a suspension with an OD450 of 1) on YEME-MOPS-agar (3 ml) and incubating the plates for 9 days aerobically with or without 50 mM nitrate. (B, C) Growth curves for the wild-type strain M145 were performed in 24-well plates in 1 ml (circles) or 2 ml (triangles) of TSB medium. The cultures whose results are shown in panel B were inoculated with spores to an OD450 of 0.25, whereas those whose results are shown in panel C were inoculated with highly dispersed mycelium (see Materials and Methods) to 150 CAE/ml. The plates were incubated without nitrate (filled symbols), with 20 mM nitrate (open symbols) (B), or with different concentrations of nitrite (filled circles and full line, no nitrite; filled circles and dashed line, 5 mM nitrite; open circles and dashed line, 25 mM nitrite; open circles and dotted line, 50 mM) (C).
The demonstration that S. coelicolor mycelial cultures can reduce nitrate anaerobically but not in the presence of oxygen initially suggested that nitrate should have only a limited effect on the aerobic growth of the bacterium in rich medium. In contrast, however, growth on yeast extract-malt extract (YEME)-MOPS-agar (Fig. 6A, right) revealed a clear retardation in the development of aerobically grown colonies of strains that could synthesize Nar2. Colonies of M145 (wild type) and NM59 (Δnar-1 Δnar-3) showed strongly retarded colony development in the presence of 50 mM nitrate, whereas colonies without nitrate sporulated normally, as indicated by a gray-white colony surface appearance and by a substantial secretion of the blue pigment actinorhodin (Fig. 6A, right). As controls, strains NM3 (Δnar-2) and NM92 (Δnar-1 Δnar-2 Δnar-3), both of which lack Nar2, exhibited a normal sporulation phenotype in the presence of 50 mM nitrate (Fig. 6A, right). Colony growth on solid medium results in local anaerobiosis, and thus, nitrite accumulates under aerobic conditions (data not shown). To confirm that the developmental retardation found on solid medium was due to the nitrate reduction, we examined the growth in liquid culture with and without nitrate (Fig. 6B). It was observed that the growth rate of the wild-type strain in small-scale liquid cultures (21) was better without nitrate than in its presence. This effect was independent of culture volume, and the same effect was observed in 1-ml and 2-ml cultures, which caused various levels of oxygen limitation. In both cases, growth was reduced by an amount equivalent to ca. 200 CAE ml−1 of mycelium (Fig. 6B). This growth limitation in the presence of nitrate was not observed in strain NM92 (Δnar-1 Δnar-2 Δnar-3) (data not shown), suggesting that the effect was potentially due to nitrite toxicity caused by Nar2-catalyzed nitrate reduction. To test this, growth curves were generated in the presence of exogenously added nitrite (Fig. 6C). Even addition of 5 mM nitrite reduced growth significantly. A larger amount of nitrite (25 and 50 mM) resulted in proportionately greater reduced growth; for each mmol of nitrite added to the culture, a reduction in growth equivalent to 22.5 CAE (21) of mycelium was observed over a period of 12 h. It is therefore likely that any growth benefit caused by nitrate reduction under hypoxic conditions is masked by the effect of the nitrite toxicity.
DISCUSSION
Early studies on nitrate respiration in vegetative stages of aerobically grown streptomycetes suggested a corespiration of nitrate and oxygen in a process termed aerobic denitrification (28, 29). The possible mechanisms and ecological implications of such a process have been controversial (28, 29), but the findings of more recent studies reported in the current literature for Microvirgula aerodenitrificans, Paracoccus denitrificans, and Pseudomonas aeruginosa suggest that the premise of the process has gained wide acceptance for Gram-negative bacteria (30, 31). In contrast to these organisms, however, S. coelicolor A3(2) is a Gram-positive bacterium without a periplasm. To date, aerobic denitrification shows a strict link with periplasmic nitrate reductases (Naps) (31). This is because Naps do not require that nitrate be transported across the membrane (32). As has been shown previously (33) and as we have demonstrated for S. coelicolor, the transport of nitrate into the cytoplasm, thus supplying substrate for the Nar enzyme (Fig. 7), by NarK-like proteins is inhibited by oxygen, and consequently, aerobic nitrate reduction for vegetative stages of streptomycetes would not be expected. Our previously observed nitrate reduction in aerobically growing cultures of S. coelicolor A3(2) (12) was actually caused by rapid oxygen consumption in cell aggregates, which resulted in localized anaerobiosis with consequent nitrate reduction. Therefore, our results agree with arguments raised in early studies that the inhomogeneity of cultures is responsible for the generation of anaerobic microniches (29) and that this is the cause of the apparent ability of Gram-positive aerobes to perform aerobic nitrate reduction. The actinobacterium M. tuberculosis has also recently been shown to reduce nitrate in human macrophages and perform NarK2-dependent nitrate reduction (34). Despite originally being considered an aerobic environment, it transpires that the human macrophage is indeed a hypoxic environment (34), and thus, M. tuberculosis, like S. coelicolor, conforms to the consensus that Gram-positive actinobacteria do not perform aerobic respiratory nitrate reduction.
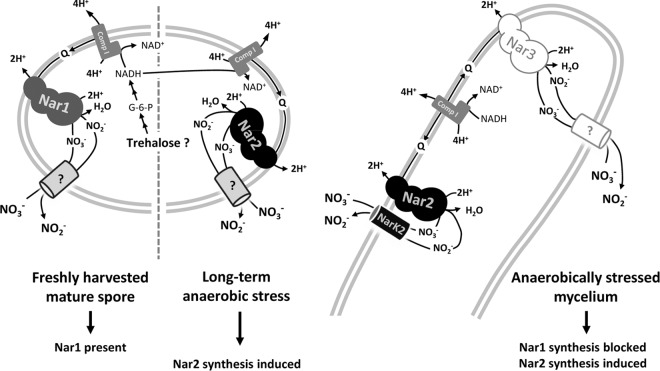
FIG 7.
Model depicting our current knowledge of nitrate reduction in spores and mycelium of Streptomyces coelicolor. Nar1 is the main Nar enzyme in spores (13). ?, the process is uncharacterized; Comp I, NADH:quinone oxidoreductase; Q, quinone.
While it has been reported that some streptomycete species carry out a nearly complete denitrification (35–37), the reduction of nitrate by S. coelicolor A3(2) is complete at nitrite excretion due to the genetically determined lack of further denitrifying enzymes in the genome (20). The NirBD nitrite reductase was shown to be functional only for aerobic nitrite assimilation, consistent with it being regulated by the global nitrogen regulator GlnR together with the coactivator NnaR (38).
Nitrite accumulation by S. coelicolor A3(2) negatively influenced growth, which has also been shown for other bacteria (39, 40). The toxic effect of nitrite presumably is a consequence of the chemical generation of nitric oxide at low pH (40). This fact seems to have been overlooked in standard Streptomyces culture medium recipes; e.g., the commonly used minimal medium HMM (41) contains 50 mM sodium nitrate as the sole nitrogen source. When growing as colonies on such minimal medium (or as cell aggregates in liquid culture), the possibility of a temporary intracellular nitrite accumulation cannot be excluded. Indeed, our studies on the differentiation of S. coelicolor A3(2) on such solid medium also demonstrated a negative influence of nitrite on sporulation, which was previously ascribed by other authors to nitrate limitation (42). The findings reported here revealed a strong developmental effect of nitrite accumulation in our rich solid medium assay and suggest that this hypothetical nitrate limitation might instead be due, at least in part, to nitrite toxicity. Exogenously added nitrite also causes growth inhibition, although whether this nitrite needs to be transported into cells to cause the toxic effects is currently unknown. It should be stressed, however, that the nitrite toxicity observed with the pure cultures and high nitrate concentrations used in this study will be unlikely to have a negative impact in the natural environment, where competing microorganism will utilize the nitrite and thus detoxify it.
Unlike in other bacteria, such as E. coli (43), neither nitrate nor nitrite appears to influence nar gene expression in either mycobacteria (44) or streptomycetes, as revealed by a proteomic study that showed high-level synthesis of Nar2 (SCO0216 to SCO0219) and NarG3 (SCO4947) in mycelium even in the absence of nitrate (45). These findings support our conclusions that hypoxia, and not nitrogen oxyanions, is mainly required for the induction of Nar2 enzyme synthesis. Notably, an abrupt shift from aerobic conditions and highly disperse mycelium to anaerobic conditions had no immediate effect on Nar2 synthesis or activity. Instead, an increase in the level of the large subunit of Nar2 was observed only after incubation under oxygen-limiting conditions. This is reminiscent of the microaerobic upregulation observed for M. tuberculosis (44), with the exception that in M. tuberculosis the synthesis of the NarK2 nitrate transporter appears to be upregulated by oxygen limitation (44) and not the Nar enzyme. The fact that low levels of NarG2 were detectable in aerobically cultivated mycelium suggests either that there was some oxygen limitation in young, germinated cultures or that developmental control also impinges on Nar2 synthesis. Further studies will be required to distinguish between these possibilities.
Nevertheless, a clear redox-dependent increase in Nar2 synthesis during hypoxia was evident. In other bacteria, such as E. coli, dual regulation is observed at the transcriptional level, whereby transcription is induced to a low level by anaerobiosis and increased further in the presence of nitrate (43). We have so far been unsuccessful in detecting an effect of nitrate on nar operon transcription in S. coelicolor; however, the transcript levels of narG2 do appear to be upregulated in transition phase. Whether this is also a consequence of a developmentally controlled process or whether oxygen limitation is involved is currently unresolved.
Freshly prepared resting spores of S. coelicolor lack detectable NarG2 polypeptide and exhibit no Nar2-dependent nitrate reduction activity, despite having narG2 mRNA transcripts. Nevertheless, long-term anaerobic incubation of spores resulted in the low-level synthesis of Nar2; however, this could be inhibited by rifampin, indicating that transcription either of the nar-2 operon itself or of another gene whose product is required for nar-2 transcript translation is responsible for Nar2 synthesis. Nevertheless, Nar2 synthesis also occurs in the presence of oxygen during spore germination and in the mycelium. Together, these data suggest that there might also be a signal that is independent of oxygen or reduction-oxidation which initiates Nar2 synthesis. The consequence of this regulation is, however, that Nar2 enzyme synthesis is essentially constitutive in mycelium in anticipation of hypoxia/anaerobiosis. Our finding that Nar2 can nevertheless be synthesized during long-term anaerobic stress of spores (see the model in Fig. 7) suggests that there must be a further regulatory mechanism controlling Nar2 synthesis. Whether this is related to the induction of Nar2 synthesis during germination is unknown. With the optimized experimental approaches developed in this study and an earlier study (21), it will be possible in the future to elucidate the regulatory control of nitrate respiration in spores (13) and in vegetative mycelium and to identify and characterize the genes and regulators that control Nar2 synthesis.
Our studies also revealed a posttranslational effect of oxygen on Nar2 activity in whole mycelium, whereby oxygen caused the immediate cessation of nitrate reduction, even in the presence of fully induced Nar2 in mycelium. Measurement of oxygen-independent nitrate reductase activity in crude extracts derived from these mycelial samples clearly demonstrates that oxygen exerts its effect at the level of nitrate transport, as has been observed for other bacteria (33, 44). Our discovery that NarK2 of S. coelicolor A3(2) appears to be specific for Nar2 (Fig. 7) and that under anaerobic conditions this results in the quantitative export of nitrite suggests that NarK2 is an oxygen-responsive nitrate:nitrite antiporter. It could be shown that NarK2 is not responsible for the oxygen-inhibited nitrate:nitrite antiport in spores (13), suggesting that each Nar enzyme might have its own specific transport protein. The transporter associated with Nar1 in spores is currently unknown. Finding a Nar-specific nitrate:nitrite antiporter also suggests a close physical association of the transporter and its cognate reductase, as suggested by the model of Nar function presented in Fig. 7. It is possible that the residual nitrite production observed in the mycelium of a narK2 mutant (Fig. 3B) results from Nar3 activity; however, it must still be demonstrated unequivocally whether nitrate reduction by Nar3 is independent of NarK2.
Our current knowledge of the roles of the three Nar enzymes in S. coelicolor is presented in the model shown in Fig. 7. The current study has demonstrated that Nar2 is the main respiratory nitrate reductase functional in mycelium. Nitrate import and nitrite export are coupled (12), and this is achieved by the activity of the Nar2-specific antiporter NarK2. Whether NarK2 synthesis is regulated in response to oxygen remains to be demonstrated. Much less is known regarding Nar3, which is active in mycelium (12). Nonetheless, it appears that substrate delivery to this enzyme is independent of NarK2. Finally, we could also show that Nar1-dependent nitrate reduction in spores is also independent of NarK2. It is conceivable that the transporter functional with Nar1 also supplies nitrate to Nar3, but future studies will be required to resolve this issue.
We have demonstrated recently that spores without an exogenous electron donor nevertheless readily reduce nitrate during anaerobiosis (13). This suggests that an internal storage compound, for example, trehalose, could supply the reducing power required to help maintain a membrane potential. It is possible that the reducing equivalents generated by the oxidation of NADH by complex I are used to reduce nitrate and thus contribute to the proton motive force (Fig. 7). A similar source of reducing power is suggested for mycelium under hypoxic or anaerobic conditions.
ACKNOWLEDGMENTS
We are grateful to Paul Dyson (University of Swansea) for supplying cosmids.
This work was supported by the Deutsche Forschungsgemeinschaft (SA 494/4-1).
Footnotes
Published ahead of print 15 September 2014
REFERENCES
- 1.Kämpfer P. 2012. Genus I. Streptomyces Waksman and Henrici 1943, 339, AL emend, Witt and Stackebrandt 1990, 370, emend. Wellington, Stackebrandt, Sanders, Wolstrup and Jorgensen 1992, 159, p 1455–1767 In Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, Ludwig W, Whitman WB. (ed) Bergey's manual of systematic bacteriology, part B, 2nd ed, vol 5 Springer, New York, NY. [Google Scholar]
- 2.Hodgson DA. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42:47–238. 10.1016/S0065-2911(00)42003-5. [DOI] [PubMed] [Google Scholar]
- 3.Niederpruem DJ, Hackett DP. 1961. Respiratory chain of Streptomyces. J. Bacteriol. 81:557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott RI, Poole RK. 1988. The carbon monoxide- and oxygen-reacting haemoproteins of Streptomyces clavuligerus: cytochrome aa3 is the predominant terminal oxidase of the respiratory chain. Arch. Microbiol. 150:465–470. 10.1007/BF00422288. [DOI] [Google Scholar]
- 5.Blundell KLIM, Wilson MT, Svistunenko DA, Vijgenboom E, Worrall JAR. 2013. Morphological development and cytochrome c oxidase activity in Streptomyces lividans are dependent on the action of a copper bound Sco protein. Open Biol. 3:120163. 10.1098/rsob.120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borodina I, Krabben P, Nielsen J. 2005. Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res. 15:820–829. 10.1101/gr.3364705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Keulen G, Alderson J, White J, Sawers RG. 2007. The obligate aerobic actinomycete Streptomyces coelicolor A3(2) survives extended periods of anaerobic stress. Environ. Microbiol. 9:3143–3149. 10.1111/j.1462-2920.2007.01433.x. [DOI] [PubMed] [Google Scholar]
- 8.Sohaskey CD. 2008. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J. Bacteriol. 190:2981–2986. 10.1128/JB.01857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham-Bussel A, Bange FC, Nathan CF. 2013. Nitrite impacts the survival of Mycobacterium tuberculosis in response to isoniazid and hydrogen peroxide. Microbiologyopen 2:901–911. 10.1002/mbo3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan MP, Sequeira P, Lin WW, Phong WY, Cliff P, Ng SH, Lee BH, Camacho L, Schnappinger D, Ehrt S, Dick T, Pethe K, Alonso S. 2010. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One 5:e13356. 10.1371/journal.pone.0013356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wayne LG, Sohaskey CD. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139–163. 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 12.Fischer M, Alderson J, van Keulen G, White J, Sawers RG. 2010. The obligate aerobe Streptomyces coelicolor A3(2) synthesizes three active respiratory nitrate reductases. Microbiology 156:3166–3179. 10.1099/mic.0.042572-0. [DOI] [PubMed] [Google Scholar]
- 13.Fischer M, Falke D, Sawers RG. 2013. A respiratory nitrate reductase active exclusively in resting spores of the obligate aerobe Streptomyces coelicolor A3(2). Mol. Microbiol. 89:1259–1273. 10.1111/mmi.12344. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 16.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin synthesis utilizing a novel integration vector. Gene 111:61–68. 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Wang Y, Yu Y, Bai T, Chen L, Liu P, Guo H, Zhu C, Tao M, Deng Z. 2012. A non-restricting and non-methylating Escherichia coli strain for DNA cloning and high-throughput conjugation to Streptomyces coelicolor. Curr. Microbiol. 64:185–190. 10.1007/s00284-011-0048-5. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Martinez LT, Del Sol R, Evans MC, Fielding S, Herron PR, Chandra G, Dyson PJ. 2011. A transposon insertion single-gene knockout library and new ordered cosmid library for the model organism Streptomyces coelicolor A3(2). Antonie Van Leeuwenhoek 99:515–522. 10.1007/s10482-010-9518-1. [DOI] [PubMed] [Google Scholar]
- 19.Gregory MA, Till R, Smith MCM. 2003. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J. Bacteriol. 185:5320–5323. 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 21.Fischer M, Sawers RG. 2013. A universally applicable and rapid method for measuring the growth of Streptomyces and other filamentous microorganisms by methylene blue adsorption-desorption. Appl. Environ. Microbiol. 79:4499–4502. 10.1128/AEM.00778-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354. 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones RW, Garland PB. 1977. Sites and specificity of the reaction of bipyridylium compounds with anaerobic respiratory enzymes of Escherichia coli. Effects of permeability barriers imposed by the cytoplasmic membrane. Biochem. J. 164:199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Cruz Ramos H, Boursier L, Moszer I, Kunst F, Danchin A, Glaser P. 1995. Anaerobic transcription activation in Bacillus subtilis: classification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 14:5984–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng H, Wisedchaisri G, Gonen T. 2013. Crystal structure of a nitrate/nitrite exchanger. Nature 497:647–651. 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson LA, Kuenen JG. 1984. Aerobic denitrification: a controversy revived. Arch. Microbiol. 139:351–354. 10.1007/BF00408378. [DOI] [Google Scholar]
- 29.Robertson LA, Kuenen JG. 1984. Aerobic denitrification—old wine in new bottles? Antonie Van Leeuwenhoek 50:525–544. 10.1007/BF02386224. [DOI] [PubMed] [Google Scholar]
- 30.Takaya N, Catalan-Sakairi MAB, Sakaguchi Y, Kato I, Zhou Z, Shoun H. 2003. Aerobic denitrifying bacteria that produce low levels of nitrous oxide. Appl. Environ. Microbiol. 69:3152–3157. 10.1128/AEM.69.6.3152-3157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carter JP, Hsaio YH, Spiro S, Richardson DJ. 1995. Soil and sediment bacteria capable of aerobic nitrate respiration. Appl. Environ. Microbiol. 61:2852–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moir JW, Wood NJ. 2001. Nitrate and nitrite transport in bacteria. Cell. Mol. Life Sci. 58:215–224. 10.1007/PL00000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez D, Rowe JJ. 1988. Oxygen inhibition of nitrate uptake is a general regulatory mechanism in nitrate respiration. J. Biol. Chem. 263:7937–7939. [PubMed] [Google Scholar]
- 34.Cunningham-Bussel A, Zhang T, Nathan CF. 2013. Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc. Natl. Acad. Sci. U. S. A. 110:E4256–E4265. 10.1073/pnas.1316894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albrecht A, Ottow JCG, Benckiser G, Sich I, Russow R. 1997. Incomplete denitrification (NO and N2O) from nitrate by Streptomyces violaceoruber and S. nitrosporeus revealed by acetylene inhibition and 15N gas chromatography-quadrupole mass spectrometry analyses. Naturwissenschaften 84:145–147. 10.1007/s001140050365. [DOI] [Google Scholar]
- 36.Shoun H, Kano M, Baba I, Takaya N, Matsuo M. 1998. Denitrification by actinomycetes and purification of dissimilatory nitrite reductase and azurin from Streptomyces thioluteus. J. Bacteriol. 180:4413–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumon Y, Sasaki Y, Kato I, Takaya N, Shoun H, Beppu T. 2002. Codenitrification and denitrification are dual metabolic pathways through which dinitrogen evolves from nitrate in Streptomyces antibioticus. J. Bacteriol. 184:2963–2968. 10.1128/JB.184.11.2963-2968.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amin R, Reuther J, Bera A, Wohlleben W, Mast Y. 2012. A novel GlnR target gene, nnaR, is involved in nitrate/nitrite assimilation in Streptomyces coelicolor. Microbiology 158:1172–1182. 10.1099/mic.0.054817-0. [DOI] [PubMed] [Google Scholar]
- 39.Rowe JJ, Yarbrough JM, Rake JB, Eagon RG. 1979. Nitrite inhibition of aerobic bacteria. Curr. Microbiol. 2:51–54. 10.1007/BF02601735. [DOI] [Google Scholar]
- 40.Dykhuizen RS, Fraser A, McKenzie H, Golden M, Leifert C, Benjamin N. 1998. Helicobacter pylori is killed by nitrite under acidic conditions. Gut 42:334–337. 10.1136/gut.42.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. 1989. Dispersed growth of Streptomyces in liquid culture. Appl. Microbiol. Biotechnol. 31:272–277. [Google Scholar]
- 42.Karandikar A, Sharples GP, Hobbs G. 1997. Differentiation of Streptomyces coelicolor A3(2) under nitrate-limited conditions. Microbiology 143:3581–3590. 10.1099/00221287-143-11-3581. [DOI] [PubMed] [Google Scholar]
- 43.Rabin RS, Stewart V. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohaskey CD, Wayne LG. 2003. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol. 185:7247–7256. 10.1128/JB.185.24.7247-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas L, Hodgson DA, Wentzel A, Nieselt K, Ellingsen TE, Moore J, Morrissey ER, Legaie R, STREAM Consortium. Wohlleben W, Rodríguez-Garcia A, Martin JF, Burroughs NJ, Wellington EMH, Smith MCM. 2012. Metabolic switches and adaptations deduced from the proteomes of Streptomyces coelicolor wild type and phoP mutant grown in batch culture. Mol. Cell. Proteomics 11:M111.013797. 10.1074/mcp.M111.013797. [DOI] [PMC free article] [PubMed] [Google Scholar]