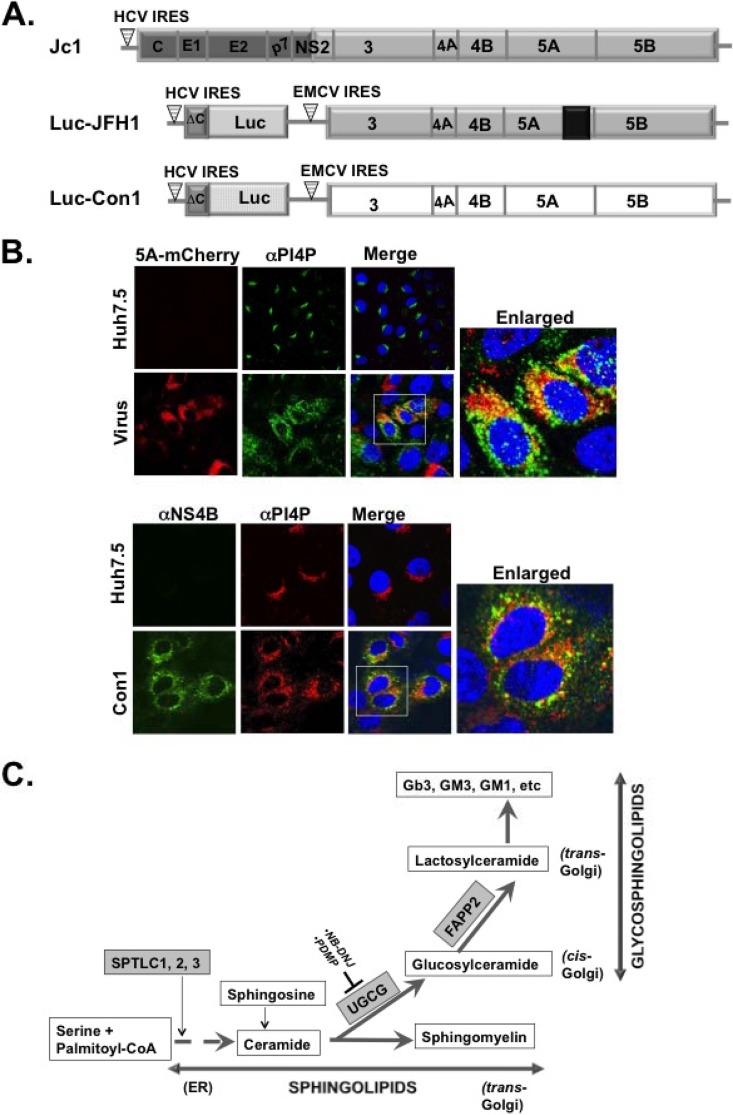
FIG 1.
(A) Schematic of the Jc1 (genotype 2a) virus and luciferase reporter replicons (Luc-JFH1 [genotype 2a] and Luc-Con1 [genotype 1b]) used to determine the role of the glycosphingolipid machinery in HCV replication. Note that NS5A has a C-terminal mCherry fusion (in Luc-JFH1 and virus used for panel B) as reported by Gottwein et al. (36). (B) Huh7.5 cells were mock infected or infected with HCV J6/JFH1 (MOI of 0.1) with a C-terminal mCherry fusion to NS5A as described for panel A (36). At 48 h postinfection, the cells were processed for confocal microscopy with mouse monoclonal αPI4P antibody (green). NS5A was detected via mCherry fluorescence. Alternatively, HCV Con1 replicon cells were grown for 48 h and stained with mouse monoclonal α-PI4P antibody (red) and rabbit polyclonal antibody against NS4B (green). The boxed areas are a magnified view for colocalization (yellow) of HCV NS5A or NS4B protein with PI4P. (C) Diagram of the de novo biosynthetic pathway leading to sphingolipids (e.g., ceramide and sphingomyelin) and glycosphingolipids (e.g., glucosylceramide and lactosylceramide) production. The SPTLC1, 2, 3 complex encodes the subunit of SPT (highlighted in gray), the first enzyme in the pathway leading to ceramide production. The SPTLC1 subunit interacts with SPTLC2 or SPTLC3 to form two distinct enzymatic functional complexes. Notice that ceramide is an intermediate product for generating both sphingolipids and glycosphingolipids. UGCG is highlighted in gray and codes for glucosylceramide synthase, a rate-limiting enzyme in glycosphingolipid synthesis. NB-DNJ and PDMP (54–59) are two pharmacological inhibitors of UGCGC. FAPP2 is highlighted in gray and carries glucosylceramide from the cis-Golgi to the trans-Golgi network for conversion into lactosylceramide and other glycosphingolipids. CoA, coenzyme A.