ABSTRACT
The gp120 portion of the envelope spike on human immunodeficiency virus type 1 (HIV-1) plays a critical role in viral entry into host cells and is a key target for the humoral immune response, and yet many structural details remain elusive. We have used cryoelectron tomography to visualize the binding of the broadly neutralizing monoclonal antibody (MAb) 447-52D to intact envelope spikes on virions of HIV-1 MN strain. Antibody 447-52D has previously been shown to bind to the tip of the V3 loop. Our results show antibody arms radiating from the sides of the gp120 protomers at a range of angles and place the antibody-bound V3 loop in an orientation that differs from that predicted by most current models but consistent with the idea that antibody binding dislodges the V3 loop from its location in the Env spike, making it flexible and disordered. These data reveal information on the position of the V3 loop and its relative flexibility and suggest that 447-52D neutralizes HIV-1 MN by capturing the V3 loop, blocking its interaction with the coreceptor and altering the structure of the envelope spike.
IMPORTANCE Antibody neutralization is one of the primary ways that the body fights infection with HIV. Because HIV is a highly mutable virus, the body must constantly produce new antibodies to counter new strains of HIV that the body itself is producing. Consequently, antibodies capable of neutralizing multiple HIV strains are comparatively few. An improved understanding of the mechanism of antibody neutralization might advance the development of immunogens. Most neutralizing antibodies target the Env glycoprotein spikes found on the virus surface. The broadly neutralizing antibody 447-52D targets the highly conserved β-turn of variable loop 3 (V3) of gp120. The importance of V3 lies in its contribution to the coreceptor binding site on the target cell. We show here that 447-52D binding to V3 converts the Env conformation from closed to open and makes the V3 loop highly flexible, implying disruption of coreceptor binding and attachment to the target cell.
INTRODUCTION
The entry of human immunodeficiency virus 1 (HIV-1) and simian immunodeficiency virus (SIV) into a target cell is initiated when the viral surface trimeric envelope glycoprotein spikes (Env), comprised of noncovalently associated heterodimers of gp120 and gp41, interact with the cell surface receptor, CD4 (1, 2). The binding of CD4 induces a conformational change in gp120 allowing HIV-1 to bind to a coreceptor (chemokine receptors CCR5 or CXCR4) expressed on the host macrophage or T-helper cell, which is followed by a structural change in the gp41 to mediate the fusion between viral and cell membranes.
HIV-1 is the most mutable virus known with different subtypes/clades expressing considerable sequence diversity, a characteristic largely responsible for the inability thus far to develop an effective vaccine (3–6). Epitopes on exposed peptide regions rapidly mutate, because of the error-prone nature of the viral reverse transcriptase, thereby pressuring the immune system to constantly produce new antibodies. In contrast, many of the structurally conserved portions of the envelope spike are masked by extensive glycosylation or are otherwise sterically occluded (7–10). Other conserved potential antibody targets are only transiently exposed during the receptor-induced conformational changes associated with the fusion process. For many years, relatively few potent broadly neutralizing monoclonal antibodies (bnMAbs), isolated from the HIV-infected individuals, have been available for study. These include 2F5, 4E10, 2G12, and b12 (11–14). More recently, a variety of additional bnMAbs have been described (15, 16).
The gp120 portion of the envelope spike (Env) is comprised of five variable regions (V1 to V5) and five constant regions (C1 to C5) (17, 18). Of these, the V3 loop plays a particularly important role serving as a significant component of the coreceptor binding site (19, 20) and as an important target for neutralizing antibodies (21–24). HIV-1 strains vary widely in their susceptibility to V3-mediated neutralization (25, 26). Neutralization resistance is presumably due, in part, to the shielding of the V3 loop by the large V1/V2 loop (27–35). One MAb with moderate neutralization breadth, 447-52D (23, 36, 37), targets the highly immunogenic β-turn at the apex of the V3 loop (Fig. 1). Structural information on the location and positional variability of the V3 loop is incomplete. The atomic structure of unliganded (38, 39) and liganded (40–42) forms of the soluble gp120 core, the gp120 core with intact V3 loop (43), and with complete N- and C-terminal peptides (44) have been published, but there are no published atomic-resolution structures of trimeric Env. Even the complete unliganded HIV-1 monomer structure has not yet been solved by crystallography. In the absence of complete atomic models of the trimeric Env, cryo-electron tomography (cryo-ET) represents the best possible choice to address a variety of important structural details.
FIG 1.
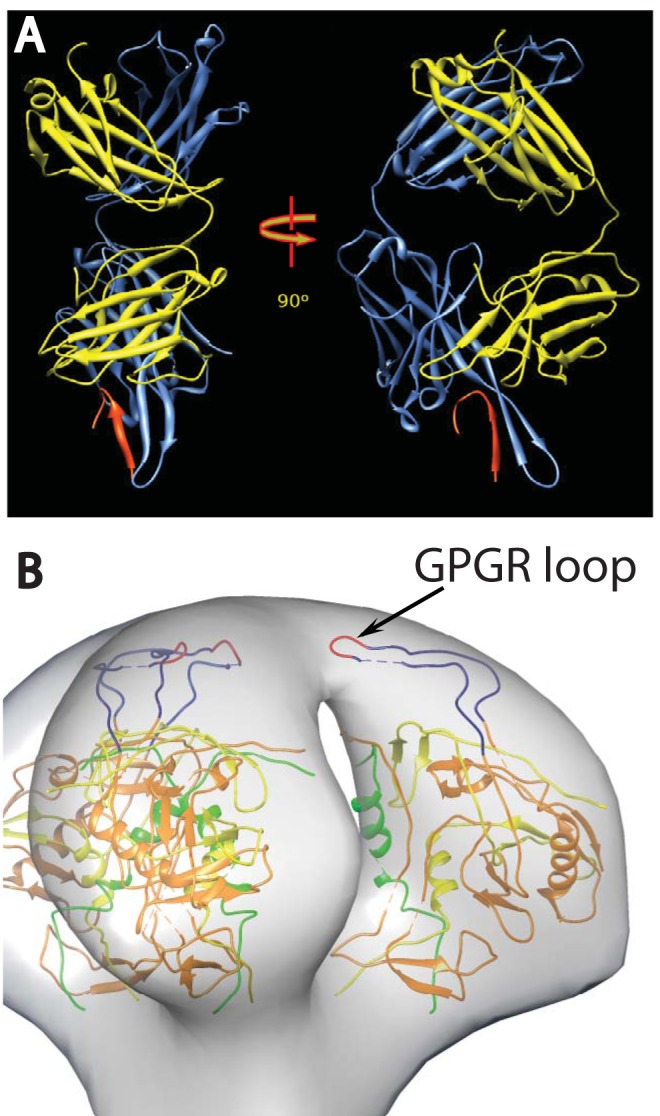
(A) Ribbon representation of the crystal structure of Fab 447D MAb from PDB 1Q1J (58). The heavy chain is colored cornflower blue and the light chain is colored yellow, with the V3 MN peptide in red. The two views are approximately perpendicular to the pseudo 2-fold axis. (B) View of a plausible spike atomic model showing the rough location of the V3 loop. The 3-fold axis of the spike is approximately vertical in the picture. The coloring scheme is as follows: gp120 residues 90 to 124, green; residues 198 to 396, orange; and residues 410 to 492, yellow. The intact V3 loop, residues 298 to 327, is navy blue, and the GPGR loop at the tip of V3 is red.
Several research groups have produced cryo-ET reconstructions showing the three-dimensional (3-D) density maps of trimeric Env on both HIV-1 and the closely related SIV (45–50). Initial studies on unliganded spikes showed them in what is now referred to as the “closed” configuration (45–47). Subsequently, in situ density maps of Env in complex with CD4, CD4 binding site (CD4bs) MAbs, and CD4 plus CD4-induced (CD4i) MAbs have been acquired, which showed that ligand binding significantly altered the spike morphology to produce an “open” configuration wherein the gp120 protomers are displayed with minimal contact to each other (50). Certain HIV-1 and SIV strains are constitutively in the open configuration in the absence of CD4 ligation, a configuration correlated with lowered resistance to antibody neutralization (48).
In an effort to help establish the location of key structural elements within the Env spike, Hu et al. (49) and White et al. (48) compared the cryo-electron microscopy (cryo-EM) density maps of HIV strains with or without V1/V2 loops. The results showed that the primary structural difference was the loss of mass at the apex of the spike. If the V1/V2 loop shields the V3 loop, then V3 is plausibly positioned toward the top of the Env spike as well (Fig. 1B). To gain further insight into the location of structural elements and assess the structural impact of the binding of broadly neutralizing MAbs, cryo-EM analysis of Env on the virion surface in complex with several MAbs has been performed (47, 48, 50, 51). We investigated here the effect of binding of the anti-V3 loop-targeting MAb, 447-52D, on Env spikes on intact HIV-1 MN strain virions and gain insights into the location and flexibility of the V3 loop.
MATERIALS AND METHODS
Viruses.
The AIDS Vaccine Program (SAIC Frederick, National Cancer Institute [NCI], Frederick, MD) supplied the highly purified aldrithiol-2-treated virus HIV-1(MN) CL.4/SUPT1, lot P3806 (HIV-1 MN). Aldrithiol-2 treatment eliminates virus infectivity while preserving Env structure and function (52, 53). The production and purification procedures of the viruses have been described elsewhere (52). MAb 447-52D was generously provided by Susan Zolla-Pazner, (NYU Langone Medical Center, New York, NY). Unconjugated 10-nm gold colloid nanoparticles were purchased from BBI Solutions, Cardiff, United Kingdom.
Sample preparation.
Samples (10 μl) of purified viral suspension (∼1.5 to 2.0 mg/ml total protein) were incubated at room temperature for 30 min in the presence of 447-52D whole antibody at a concentration corresponding to an estimated 10-fold molar excess over Env trimers. The sample was then mixed with 10-nm colloidal gold (used for better tracking during tilt series collection), and a 3.5-μl sample was placed on a 200 mesh R2/1 Quantifoil grid (Quantifoil Micro Tools, GmbH, Jena, Germany). Excess liquid was blotted with filter paper from both sides of the grid to form a thin layer of virus-containing buffer, which was then rapidly frozen by plunging the grid into a liquid/solid ethane slush (about −180°C) using a home-built freezing device at 25°C. The grids were either used immediately for EM examination or stored in liquid nitrogen for later use.
Cryo-EM. (i) Data collection.
HIV-1 MN frozen virus specimens were examined at liquid nitrogen temperatures under low dose conditions using a FEI G2 Polara electron microscope (FEI) equipped with a field emission gun and WAC 4k×4k charge-coupled device camera (TVIPS, Gauting, Germany). The microscope was operated at 300 kV and a magnification of ×31,000, resulting in an effective pixel size of 0.58 nm. Single-axis tilt series were collected using the FEI batch tomography capability of the Xplor3D software at a defocus of 5 μm with a cumulative dose of ∼100 e-/Å2. The angular range of the tilt series was from −65° to +65° and consisted of 65 images recorded at fixed tilt increments of 2°. The tomograms suggested that the specimen thickness was ∼200 to 250 nm.
(ii) Image processing.
The tilt series were aligned based on the intrinsic features of the individual projection images and not on the gold fiducial markers. Tomograms were computed by weighted back-projection as implemented in the Protomo software package (54). The fiducial gold particles were masked out before alignment and replaced by densities having the mean value of the whole image (55). For HIV-1, 30 tilt series were used consisting of 285 virions from which 3,618 spikes were selected manually for subtomogram alignment, classification, and averaging. Spike densities found to be much smaller than the expected size of the trimer were excluded.
With the assumption that the vector (here defined as the polar vector) from the center of the virus through the center of the spike is oriented perpendicular to the viral surface, the initial Euler angles for the polar vector were determined by a least-squares fitting routine (55). Spike subtomograms were then aligned by translation alone, and a global average was generated, which served as an initial reference to align first the polar vector and then the spin angle, which brings out the trimeric structure. In the ensemble of aligned Env spikes there is no region in the Fourier transform of the global average and the class averages that has poor data coverage due to the missing wedge effect since the spikes are uniformly distributed in angular orientation. From the aligned raw spikes, 10 class averages were computed using multivariate data analysis and hierarchical ascendant classification (55, 56). After only a few cycles of refinement the class averages clearly showed the inherent 3-fold symmetry in the spike structure. At each cycle, ca. 50 to 60% of the classes showing the clearest features in all regions of the spike were selected and used as references for the next cycle. This procedure was repeated 15 times until the appearance of the class averages no longer changed. The spike alignment mask included the spike and a small portion (outer leaflet) of the membrane, whereas the classification mask included only the spike and excluded the membrane. Previous work has shown that including the density corresponding to the lipid bilayer in the classification introduces a bias that separates spikes extending out the sides of the virions, where the membrane densities are prominent, from those that extend out the top, where the bilayer is not visible or has low contrast (55).
The global average of antibody-bound Env spikes showed only a small additional density contributed by antibody (see Movie S1 in the supplemental material), suggesting that the antibody binding sites were not saturated, the antigen-antibody complex was disordered or both. In addition, the initial classification using the entire Env spike indicated partial occupancy in that significant extra density was not apparent on one or two arms in many cases and that where additional density from the 447-52D antibody was observed, it showed considerable variation (see Movie S1 in the supplemental material). Therefore, we devised a strategy to reduce the complexity in the antibody-binding pattern. Disregarding disorder, there are eight possible arrangements in which 0 to 3 antigen sites can be bound by antibody in the population of aligned Env spikes. The antigen sites on the Env spike can be completely saturated with antibody or be antibody free. In addition, there are three ways of arranging two antibodies per spike and three additional ways of arranging a single antibody/spike. If all spike arms were superimposed on a single site rather than distributed over three sites, we could reduce the complexity in the antibody labeling by 4-fold to two possible patterns: antibody bound and antibody free. Conformational heterogeneity would then be expressed in classes showing density due to bound antibody. This strategy also increases 3-fold the number of subvolumes being averaged. Thus, after aligning all Env spikes and centering them on the 3-fold axis, we created two additional copies of each raw spike, one rotated 120° about the spike axis and the other 240°, so that each arm of the Env trimer was superimposed on a single site. We then performed a classification using a mask that included only the volume around a single spike arm. One of the ten class averages lacked density due to antibody (see Fig. 3A), while the other nine showed density configurations of various shapes.
FIG 3.
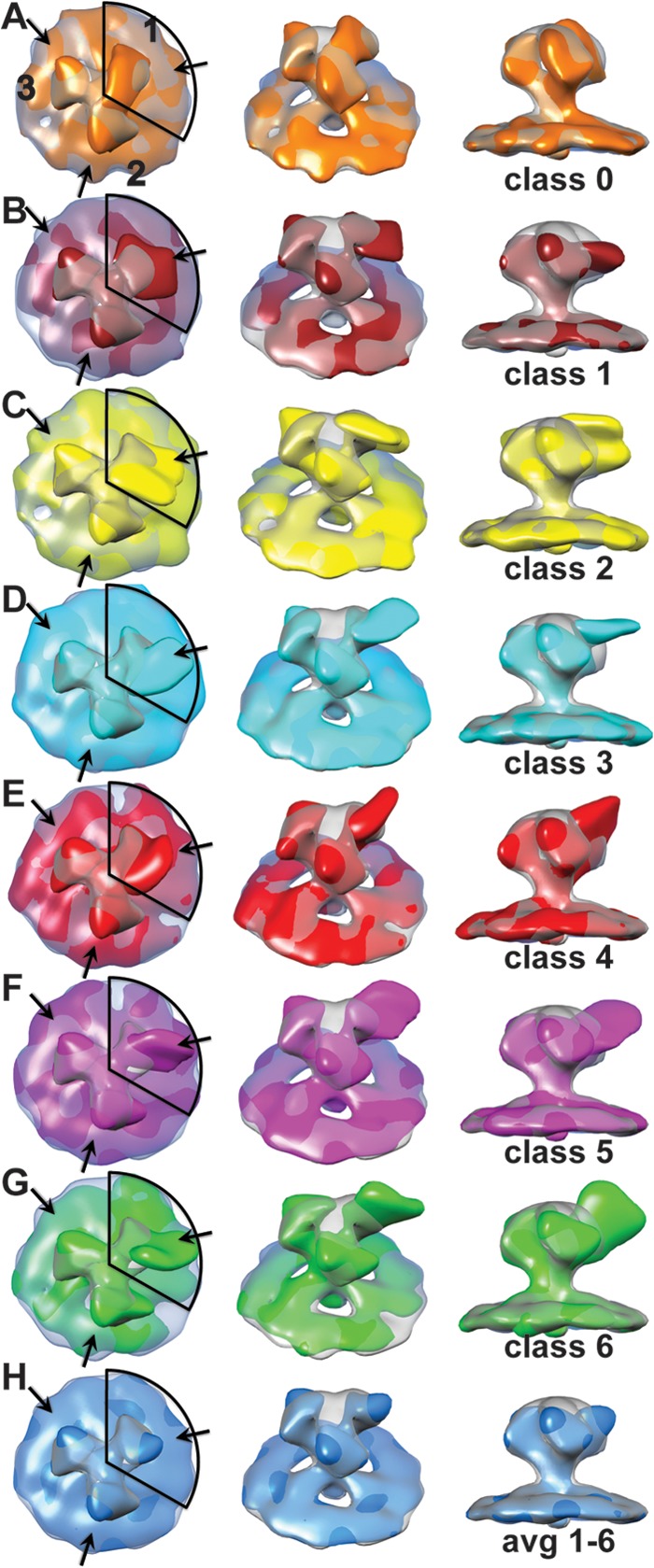
Density maps from the one-arm classification of HIV-1 MN Env spike with bound 447-52D antibody. In the left-hand column is a view down the 3-fold axis of the spike. The numbers in the top row of column A label the spike arms, thereby defining each arm as to whether classification was carried out, arm 1, or not, arms 2 and 3. The central column is a view obtained by rotating the spike average by 45° about the horizontal axis and away from the observer. The right-hand column is a view tangential to the membrane plane. Each map is superimposed on the symmetrized average of the unliganded control spike arm at 50% transparency. The contour threshold used for the unliganded control spike is the same for all figures that show it. (A) The class average having the least density due to bound MAb, which was used for making the symmetrized control spike. (B to G) The six class averages showing density due to bound antibody. (H) Average of classes 1 to 6 weighted according to the number of class members. The coloring scheme is as follows: class 0, orange; class 1, brick; class 2, yellow; class 3, cyan; class 4, red; class 5 magenta; class 6, green; and the average of 1 to 6, dodger blue. These images can also be seen as an aligned, animated, and annotated sequence in Movie S2 in the supplemental material.
The symmetrical trimer used as a “control” spike was generated by taking the class average, which had no apparent antibody density, i.e., class 0, masking it with a 120° circular segment mask and rotating the masked class 0 by 120° and 240°. The three spike arms were then summed together and low pass filtered to produce a spike with 3-fold symmetry.
Choice of contour threshold.
With the exception of the difference maps, the contour threshold for interpreting and displaying each of the different averages was chosen as follows. The contour threshold for the unliganded spike average was chosen to surround the atomic model generously, because the atomic model is incomplete. Once selected, this threshold was imposed on all figures showing the unliganded spike average. This threshold generally shows density across the spike tip. Thus chosen, the contour threshold for all global and class averages, either from the whole spike classifications or the one-arm classifications, was adjusted so that those parts of the spike not bound by antibody, which includes the counterclockwise side of the spike arm and the membrane, were contained within the boundary of the unliganded spike average. To the extent that the shapes did not match, a compromise between being completely contained and extending outside the unliganded spike was made subjectively. Although this method of selecting the threshold was somewhat subjective, it produced a consistent result because raising the threshold of the unliganded control spike, thereby reducing the density at the peak and the volume of the spike arms, also resulted in the threshold for all the averages being raised to meet the criterion.
Generation of difference maps.
The significance of the difference in density between native Env spikes and antibody-bound spikes was obtained by applying to both maps a cylindrical mask that excluded the membrane densities and surrounding background regions at a distance beyond which the bound Fab arms would be expected to extend. The maps were low pass filtered to 3.8-nm resolution, which was the Fourier shell correlation result obtained from the global average. Because each class average has fewer spikes than the global average, all of the class averages have lower resolution than 3.8 nm. The maps were normalized by subtracting the mean value and subsequently dividing by the root mean square density. The difference between the two maps was calculated using the script “get_diffmap.py,” which used functionality within the EMAN2 software package (57).
Atomic model building.
The atomic models used for fitting purposes included the atomic gp120 construct, Protein Data Bank (PDB) identification number 3DNN, derived from the association of three atomic monomer subunits as described by Liu et al. (47). The V3 loop from PDB 2B4C (41) and the 447-52D Fab complex (58) from PDB 1Q1J were added into the 3DNN structure as previously described (49) to facilitate placement into the antibody-bound density map. The atomic models were fitted into the density map using the normal mode flexible fitting (NMFF) procedure (59) obtained from the MMTSB tool set (60). The gp120 model fitted in such a way that the V3 loop and the V1/V2 stump were oriented toward the top of the spike and the N and C termini toward the bottom similar to the fitting in our previous work (49). Density maps and atomic models in all the figures were displayed using CHIMERA (61).
RESULTS AND DISCUSSION
The trimeric Env on HIV-1 and SIV surfaces define the viral tropism, mediate the fusion process, and are the prime target of the humoral response (62). Therefore, Env is the first choice as a vaccine candidate. However, despite intensive efforts, the structural and functional details of Env that may be needed to facilitate these efforts are not yet fully defined. Recent cryo-EM structures in combination with data derived from X-ray crystallography of incomplete soluble monomeric Env subunits have revealed details of the unliganded Env spike structure, as well as spikes liganded with CD4 and CD4bs and CD4i MAbs (47–51, 63, 64). For example, interaction with soluble CD4 and CD4i MAb 17b, which binds near the base of the V3 loop, triggers a conformational change from a closed configuration to an open configuration (47). In contrast, bnMAb b12 induces a partially open gp120 rearrangement, and bnCD4bs MAb VRC01 appears to exert its neutralizing effect by locking gp120 in the closed configuration (64). Consensus is building on the likely positions and functional interactions of many of the major structural components, including the variable loops (47–49).
The V1/V2 loop, which plays an important role in stabilizing the trimer and in shielding the chemokine receptor binding site prior to CD4 triggering, appears to emanate from the core of the gp120 protomers arching upward to meet the apex of the spike (48, 49). The V3 loop plays an important role in the function of the Env spike by serving as part of the coreceptor binding site following CD4 ligation (20, 65) and provides a key target of the humoral immunity (66–68). The crystal structure of the CD4-liganded gp120 core with an intact V3 loop shows the loop protruding about 30 Å above the core and, presumably, toward the target cell (58). Most data and models place the distal part of the V3 loop in close proximity to, and likely masked by, the V1/V2 loop (9, 69) to be exposed and extended upon CD4 binding (1, 70).
Human MAb 447-52D, first isolated as a heterohybridoma derived from an HIV-1 infected patient (71, 72), binds to the V3 variable loop of the HIV-1 gp120 and exhibits significant neutralizing breadth, mostly within clade B (23, 73, 74). The core epitope of the V3 sequence was mapped by immunochemical studies to the highly conserved GPGR motif in the crown of the clade B V3 loop (75–77). T-lymphoid cell line-adapted HIV-1 strains, as well as some primary isolates that express this sequence motif, are efficiently neutralized by 447-52D (23, 71, 78). The atomic structure of 447-52D in complex with V3 peptides shows that the antibody binds to the distal tip such that the pseudo 2-fold long axis of the Fab arm and the extended V3 loop are in rough alignment, i.e., the Fab arm protrudes from the tip of the V3 loop as shown in Fig. 1A (58). It is worth noting that the sequence of the V3 crown is fairly well conserved suggesting structural conservation as well. Because several different elements spread over the surface of the crown are targeted by other broadly neutralizing MAbs (79), it seems likely that the crown can be quite exposed.
To gain additional insights regarding the placement of the V3 loop in the native unliganded Env spike and to investigate the nature of anti-V3-mediated viral neutralization, we generated and analyzed cryo-ET images of virions in complex with MAb 447-52D and compared them to the structure of unliganded Env. We chose the HIV-1 MN/SUPT1 because it shows particular sensitivity to V3 neutralizing antibodies (80). The affinity of antibody 447-52D for HIV-1 MN has been measured in vitro using surface plasmon resonance of glycosylated, recombinant gp120 and shown to be 2.1 nM (81). The dissociation rate constant of 34 × 10−5/s measured under conditions that favor single Fab arm binding suggests that dissociation is insignificant under our conditions. Differences in the conditions used in that study and ours make precise predictions difficult. Nevertheless, it is worth pointing out that saturation was generally achieved in <30 min with concentrations ∼1/30 of those used here. VanCott et al. (81) also found that a concentration of 0.2 μg/ml, a value 800-fold lower than that used here, was sufficient for 50% viral neutralization, although the incubation time was longer. Thus, our conditions for antibody labeling were expected to produce high binding of antigen sites.
Representative cryo-ET images of HIV-1 MN/SUPT1 in complex with MAb 447-52D (Fig. 2A) show clearly resolved spikes in about the same number as previously reported (average of 8 to 10 spikes/virion) (82, 83). Although associated MAbs were less obvious in most of the virions, some virions showed spikes with attached antibody densities (Fig. 2B to E) when the original tomograms underwent noise reduction. Figure 2D shows a virion with spikes on opposite sides. In most cases, Env spikes were somewhat clustered on the surface of the virion, as observed previously, rather than randomly dispersed (45, 84).
FIG 2.
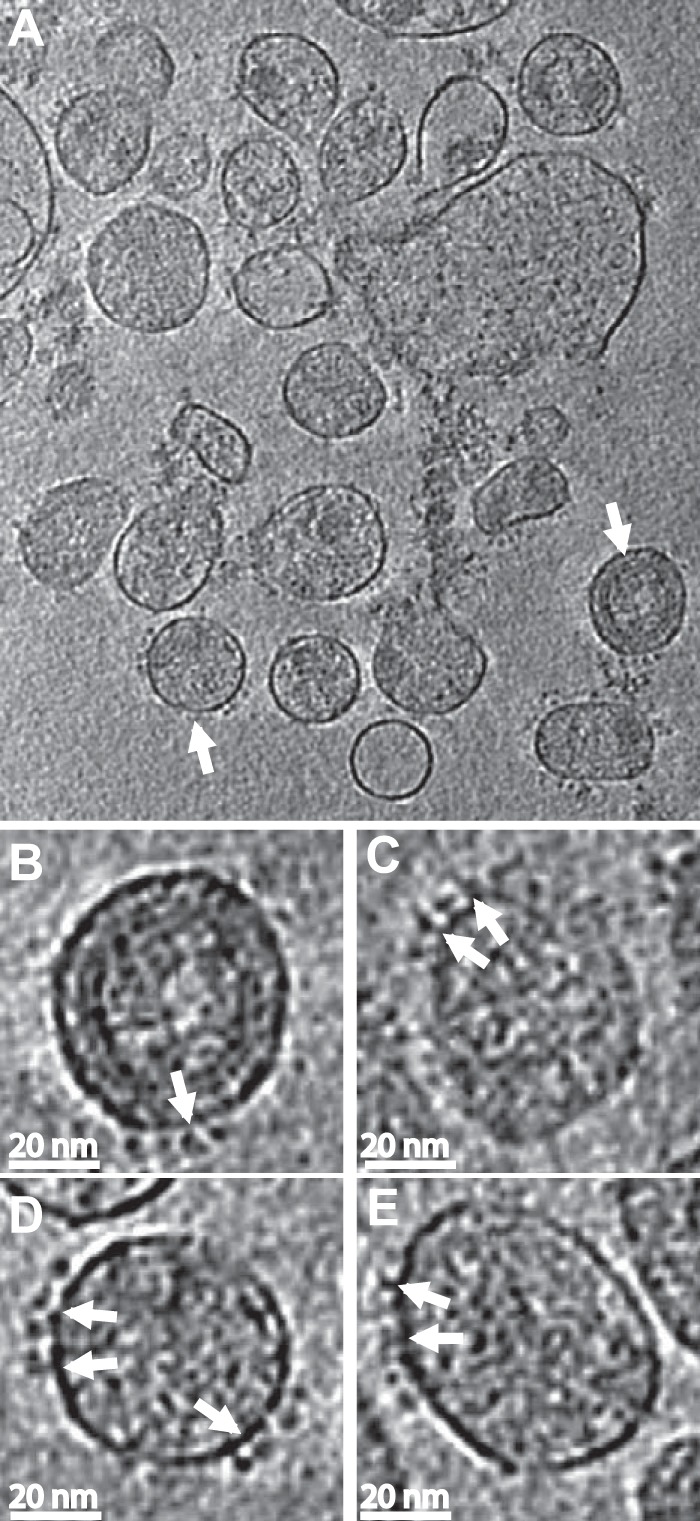
Images of antibody labeled HIV-1 MN virions. (A) A slice through the x-y plane of one of the HIV-1 MN cryo-electron tomograms. The spikes are significantly fewer compared to the short-tailed SIV (it has more spikes on its membrane surface due to truncation of the cytoplasmic tail) but still visible on the viral surface. Parts of the cores within some of the virions are also visible but the AZT treatment significantly disrupts the core of the virion. Scale bar, 100 nm. (B to E) Representative slices through individual virions following binning. The density range displayed is the mean ± 3σ. Binning (averaging the densities of a 2×2×2 cube into a single voxel) enhances the signal-to-noise ratio, thereby improving the visibility of the spikes and bound antibody. The tomograms were then reinterpolated back to the original voxel size to reduce pixelation. Panels B and D come from virions marked with white arrows in the lower-magnification view shown in panel A. The binding can be seen clearly in some of the spikes (white arrows) on the viral membrane. Antibodies sometimes extend up relative to the membrane plane and sometimes parallel to the membrane plane.
We initially carried out a classification using a mask that encompassed all three spike arms, which resulted in most classes showing less than full saturation and high variability in the density that could be assigned to antibody (see Movie S1 in the supplemental material). One of the ten classes generated showed evidence of V3 ligand saturation, five classes showed evidence of two MAbs bound, and the other four showed evidence of a single MAb bound. From the number of liganded sites and class members, we compute 56% percent antibody saturation. This could be an underestimate because some classes might have low occupancy on some spike arms and thus appear unbound by antibody. Compared with a “control” spike lacking MAb density (see below), all of the class averages show weak density at the peak in the center of the spike. Because cryo-ET data are inherently noisy, clear examples of individual spikes with three distinct MAbs binding were not obvious upon visual inspection of the class members; hence, we cannot directly assess the degree to which the three epitopes on the spikes are saturated.
Multivariate data analysis as applied here to group spike subvolumes according to similarity is more accurate when the complexity of the patterns it is attempting to analyze is simpler. We therefore sought a strategy that might simplify the complexity caused by incomplete epitope saturation and conformational heterogeneity due to inherent or induced V3 flexibility as described in Materials and Methods. We used the single-arm classification procedure to compute 10 class averages, from which we identified one class containing ∼1,100 spike arms that had no MAb bound (Fig. 3A; see also Movie S2 in the supplemental material). This class average differed from the other nine not only in the density that projected out from what appeared to be the antibody binding site based on the other classes but also in the presence of substantial density bridging Env spike arm 1 with arm 3. This density was completely lacking between arms 2 and 3 and between arms 2 and 1 (moving clockwise around the spike).
We used this class to compute a 3-fold symmetric “control” Env spike lacking MAb (most easily seen in Fig. 4A). Although this control spike was obtained unconventionally, it has the advantage of having exactly the same quality as the class averages with regard to resolution, specimen thickness, defocus, and alignment errors. Consistent with the simplicity achieved, our saturation of binding sites in this classification was ∼90%, i.e., not full saturation but certainly high enough to reveal the effect of the MAb binding and more consistent with the affinity of 447-52D for HIV-1 MN.
FIG 4.
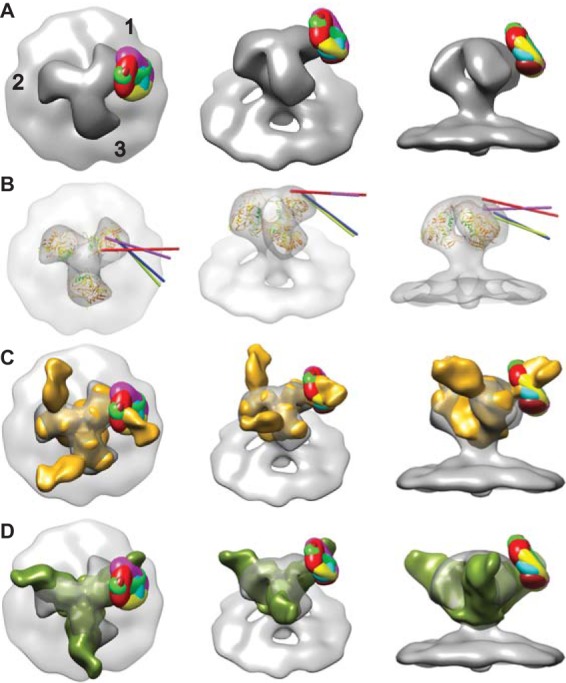
(A) Superimposed difference maps of all antibody-bound class averages with the symmetrized, antibody-free average. From left to right top view; 45° oblique view; side view tangential to the membrane. The numbers in panel A label the spike arms, thereby defining each arm by whether classification was carried out, arm 1, or not, arms 2 and 3. Images of the individual difference maps overlaid on the control spike can be visualized in Movie S3 in the supplemental material. (B) Intersection of the vectors through the quasi 2-fold axis of the Fab atomic model fitted by NMFF to the class averages with the symmetrized, antibody-free spike envelope. These four class averages are the only ones for which the vector intersected. Each of the four vectors intersects the envelope approximately at the location of the base of the V3 loop. (C) Comparison of difference maps from HIV/447D Env spikes with the soluble, partially deglycosylated spike trimer KNH1144 SOSIP 664G with antibody PGT128 bound (colored goldenrod), EMDB code emd-1970 (89). Images of the individual difference maps overlaid on emd-1970 can be seen in Movie S5 in the supplemental material. (D) Comparison of the difference maps for the 447-52D class averages with the 3-D reconstruction of PGT135 in complex with the BG505 SOSIP.664 gp140 trimer (90) colored olive (EMDB code emd-2331). Images of the individual difference maps overlaid with emd-2331 can be seen in Movie S6 in the supplemental material.
The remaining nine classes had additional density of various shapes consistent with MAb binding. Six of the nine showed very clear pairwise similarity. The hierarchical ascendant classification method used (56) provides information on the relationships among the different classes, and from this we deduced that these pairs could be combined to give a total of six MAb bound class averages. These six class averages show additional density having different shapes on arm 1 but consistent with a variable conformation of Fab (Fig. 3B to G). A small additional density in the same location as that on arm 1 is found on arms 2 and 3. The weighted average of these six classes (Fig. 3H) shows a small additional density due to MAb binding on arm 1 and a somewhat smaller density on the two arms that were not included within the classification mask as expected. Moreover, the density in the top center of the Env spike, as well as the density in the connecting adjacent arms, is low.
The one-arm classification used here specifically only looks for density patterns on the spike arm that falls within the mask and clusters the raw spike subtomograms accordingly. The averages, however, are averages of complete spikes. At 90% labeling with antibody, one might expect to see some antibody density on the other two spikes, but one-arm classification would not produce significant density there unless there was coupling between the Fab position on the arm used for classification (arm 1) with that on the other two spike arms. In all of the six single-arm classes, the two arms that were not used in the classification (arms 2 and 3) appear to be very similar, both across classes and within classes and show only a small additional density clockwise of the spike arm on the periphery, which has a shape very similar to the weighted average of the six liganded spike arm class averages (Fig. 3H). The reduction in size compared to the added density on arm 1 can be explained by the presence of unliganded spike arms in positions 2 and 3 (because these are averages over whole spikes), while unliganded spike arms are specifically excluded from the class averages on arm 1.
It should be noted that the classification method used here does not necessarily define each class as a distinct MAb protomer conformation, especially in situations that exhibit considerable flexibility. Rather, the method will bin a continuum of conformations into a specified number of classes according to the highest similarity. The number of classes specified reflects a trade-off between signal-to-noise improvement (fewer classes but more members per class) and the identification of the highest number of possible distinct conformers present (fewer members per class, noisier and less-interpretable data). Although whole antibody was used for this study, during the classification process we used a mask of a size that extended outward from the spike surface far enough to capture liganded Fab fragments. Because of antibody hinge flexibility, Fc and unassociated Fab density would be diffusely distributed in the averages.
To further assess the location(s) of the crown of the V3 loop following 447-52D liganding, positive difference maps were generated by comparing the normalized density maps of the symmetrized control MAb-free spike to those with bound MAb. Collectively, they show significant density differences (see Movie S3 in the supplemental material) at the outer periphery on the clockwise side of the spike arm, a pattern consistent with a visual comparison of the density maps of spikes displaying unbound and antibody-bound spike classes. To assess the degree of variation in Fab arm impingement on the gp120 protomers between classes, we superimposed the all six difference maps onto the symmetrized, antibody-free class average (Fig. 4A). The difference peaks all cluster around a single location on the surface of gp120 near the base of the V3 loop of the closed conformation atomic model. If the class averages represent strong binding of the antibody to a flexible antigen, then the peak of added density is expected to be close to the pivot of the motion, in this case near the base of the V3 loop.
Because the V3 loop, in particular the GPGR motif within the antibody binding site (Fig. 1B), has been proposed to contribute to the upwardly projecting arches in Env, we had expected the Fab arms to be bound closer to the apex of the spike if the spikes are untriggered and in the closed configuration (48, 50). However, the 447-52D Fabs were seen to project more radially than anticipated, i.e., they projected from the periphery of the spike arm rather than the apex. There are several possible explanations for this configuration. For example, perhaps there is a certain amount of structural “breathing” during which the V3 loop, or at least its tip, occasionally samples the more exposed lateral space and, upon 447-52D binding, is prevented from returning to its original position. Alternatively, the epitope may be positioned more toward the spike apex, and there exposed but Ab-induced conformational changes may alter its position so that it extends outward from the V3 base. Finally, in contrast to prevailing models, the V3 tip may already rest in the indicated radial position in the unliganded state but be exposed by transient changes in other structural elements whereupon antibody binding locks in the transient changes.
When the contour threshold of the class averages is chosen to match the outer parts of the second and third spike arms with the control, MAb-free spike, the match is excellent, especially counterclockwise from the V3 loop base and within the membrane density. This results in little connectivity across the top of the spike or between spike arms, where V3 and V1/V2 are believed to be located. This may seem counterintuitive but is explicable by the way the averages are generated. Although the classification is done on a single arm, the class averages are generated over complete Env spikes. With 90% antibody labeling, we expect the density on spike arms 2 and 3 to be dominated by disordered MAb, as well as any structural changes of the spikes themselves. The <10% unliganded spike arms present on arms 2 and 3 would also dilute these changes. Thus, the additional, disordered density contributed by MAb bound to arms 2 and 3 in class averages 1 to 6 is very similar to the added density seen on the same arms in the average of classes 1 to 6. Although the antibody may be disordered leaving only a small residual density, other changes in the spike arm would still be retained as long as they are ordered. These would include removal of V3 from its association with V1/V2 and consequent possible disordering of V1/V2.
If the position of the V3 loop in the unliganded Env spike is toward the top and clockwise of the spike arm, as depicted (Fig. 1B) in the neighborhood of the V1/V2 loop with which it is known to interact (27–35) and antibody binding is associated with a change in its position, then expectations are for a small reduction of mass where the V3 loop was originally positioned. Although the antibody adds a very large positive mass to the spike, removal of V3 from the peak removes comparatively little density from the center of the spike, most likely too little to detect unless it were coupled to changes in V1/V2 such as disordering. Negative peaks in the difference maps were not informative because they are dominated by differences in the region of the MAb toward the periphery of the spike, although there is a small negative peak near the base of V1/V2. For this reason, we chose instead to select contour thresholds for display systematically as described in Materials and Methods. All of the six liganded one-arm class averages, the ten three-arm class averages, and their global averages appear to have less density in the center of the spike, as well as connecting the arms than would be explicable by V3 removal alone.
This loss of density toward the apex does not appear to be due to a shift in the arm position for two reasons. First, the counterclockwise side of each spike arm in the averages has nearly the same shape as the unliganded spike arm. Differences occur primarily on the clockwise side of the arm, where the antibody is bound. Second, the radius of the centers of mass of each gp120 atomic model fit to both antibody-free and antibody-bound spike arms showed at most a 0.2-nm change, an insignificant amount at the resolution of our data. The appearance toward the apex, as well as between spike arms in the three-arm class averages as well as the one-arm, liganded spike classes, i.e., classes 1 to 6, as well as arms 2 and 3 of the unliganded class average, i.e., class 0, are very characteristic of the V1/V2 deletion mutants (48, 49). Only arm 1 of the unliganded class average (class 0) departs from this trend. Our data therefore indicate that 447-52D binding changes the spike conformation in a way consistent with a closed to open configuration. Spike arms that remain unliganded appear more or less unaffected by the changes due to 447-52D binding in their neighbors.
Since functional Env spikes on HIV-1 particles are few in number (45, 47, 82, 85), it has been suggested that cross-linking of adjacent spikes with the concomitant avidity enhancement may not play a significant role in HIV-1 neutralization (86) (evidence of spike clustering [45, 84] would, however, argue against this point). Moreover, because of the radial arrangement of potential epitopes on the trimeric spikes and the heavy glycosylation of the crown, subunit cross-linking within a given spike by most anti-Env antibodies would be unlikely (87). Consequently, from the perspective of the virus, limiting the scope of antibodies capable of neutralizing to those with high-affinity single-arm binding capability may be an evolutionary adaptation to evade humoral immunity (86). In line with this conjecture, our visual inspection of the tomograms did not indicate the presence of clearly defined examples of cross-linking of Env spikes by 447-52D on virion surfaces (data not shown). In addition, among the three-arm class averages, only one, class 1 (see Movie S1 in the supplemental material), showed density that appeared to bridge a pair of V3 sites. This class contained only 380 out of a total of 3,618 spikes or a frequency of ∼10% of the time. Although our atomic model for the unliganded spike, as well as most competing models of the spike arm, places the 447-52D epitopes in close proximity at the apex in the untriggered virus, this does not seem to lead to a high frequency of spike arm cross-linking as might be expected. It would seem more likely that antibody binding occurs with the V3 loop otherwise placed unfavorably for intraspike cross-linking. Thus, if this interpretation is correct, intraspike antigen cross-linking by 447-52D rarely if ever happens.
To better understand the V3 loop conformation and the basis for neutralization by anti-V3 MAbs, we attempted fitting of relevant atomic structures into our density maps. Flexible fitting at this resolution is not an exact process but has the value of being objective. Manual fitting is also not an exact process but has the disadvantage of complete subjectivity. Anti-V3 MAb 447-52D in complex with HIV-1 MN V3 peptide has been crystallized (58, 77, 88), as has a gp120 core structure with an intact V3 loop (43). We first aligned the atomic structure of gp120 that contains an intact V3 loop (PDB 2B4C) with the atomic gp120 trimer construct (PDB 3DNN), which is a closed conformation model derived from cryo-EM. We then aligned the crystal structure of 447-52D Fab in complex with a 16-residue MN V3 peptide (PDB 1Q1J) (58) with the V3 loop of PDB 2B4C to generate a gp120 structure with a complete V3 loop and an attached 447-52D Fab, as shown in Movie S4 in the supplemental material. The V3 loop, with associated Fab, was independently adjusted using normal mode flexible fitting to fit the various densities generated by the associated Fabs observed into six of the class averages. The results, shown in Movie S4 in the supplemental material, are consistent with flexibility within the V3 loop when bound to MAb 447-52D.
A comparison of classes where the pseudo 2-fold axis of the Fab from the NMFF fitting intersects the symmetrized, unliganded spike average shows that the 447-52D Fabs (Fig. 4B) vary in their orientation with respect to the cognate gp120 subunits to which they are attached, suggesting an inherent flexibility in the V3 loop, at least when bound with this specific MAb (Fig. 4A and B). In cases where the pseudo 2-fold axes intersect the unliganded, control spike envelope, they do so near the base of the V3 loop in the atomic model. Since each class average likely represents binning within a continuum of individual spike images rather than distinct alternative configurations, the individual Fab moieties likely vary in geometry from one to another. However, the limited resolution at the individual spike level does not permit a more detailed assessment.
We also compared the position of our difference peaks with recently published 3-D reconstructions of a soluble, partially deglycosylated Env spike complexed with anti-peptidoglycan MAbs PGT128 (89) and PGT135 (90). The soluble Env spike is a truncated version of gp120 lacking the V1/V2 loop (among other elements) and expressing a truncated V3 loop. Both reconstructions were obtained from images of negatively stained specimens using single-particle reconstruction methods. The two reconstructions were not of equal quality; the PGT128-spike complex having a better defined Fab density than the PGT135-spike map. The reconstruction of the PGT128-spike complex could be readily aligned with our unliganded reference reconstruction and compared to the difference peaks. The comparison showed that the PGT128 Fab bound nearly in the same location as the 447-52D difference peaks (Fig. 4C; see Movie S5 in the supplemental material).
We also fit the crystal structure of the PGT128 antibody Fab bound to an expressed outer domain of gp120 with a shortened V3 loop into the negative stain reconstruction and compared it to our starting antibody-free atomic model and our unliganded Env spike density map. The comparison showed that the atomic models were off radially and axially by about the diameter of an α-helix and with a slight rotation (Fig. 5). This might be explicable for several reasons. The soluble Env construct lacks the V1/V2 loop, which would be positioned in the apex of the spike, and whose absence may result in an inward shift of the gp120 spike arm. It may also be due to differences in actual magnification, or it could indicate inaccuracy in our unliganded spike atomic model.
FIG 5.
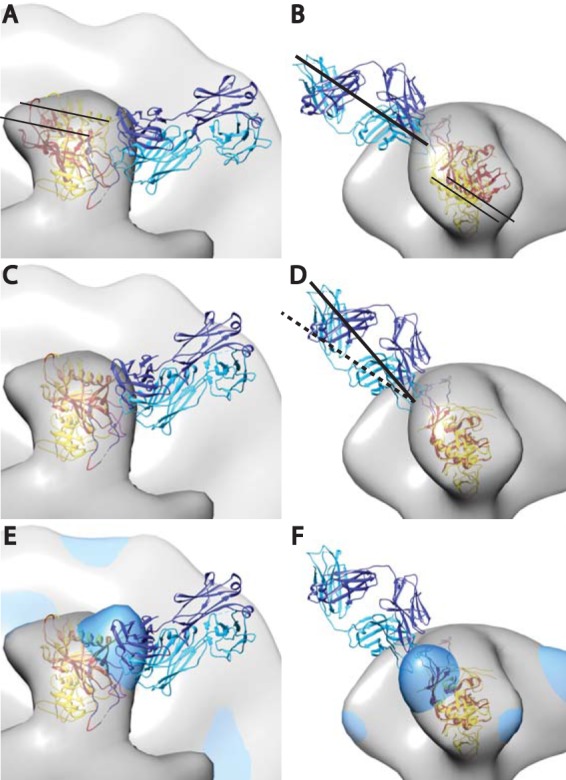
Crystal structure of PGT128 Fab in complex with an engineered glycosylated gp120 outer domain with a miniV3 loop (eODmV3), PDB 3TYG (1), aligned onto the gp120 spike pseudoatomic model from the present study. The 3TYG crystal structure is shown in orange-red, and the gp120 pseudoatomic model from the present work shown in yellow. The PGT128 Fab is shown in blue and cyan. The symmetrized average Env spike from the present study is provided as a point of reference. Panels A and B show the crystal structure as it would be positioned when fitted in the PGT128 image reconstruction (emd-1970), followed by alignment of emd-1970 to our control spike reconstruction. Panels C and D are after alignment on the gp120 atomic model used in the present study. The alignment was done manually using CHIMERA. The major movement requires a small outward radial movement and a small downward axial movement as indicated by the narrow black lines. There is also an ∼15° clockwise rotation along an axis roughly perpendicular to the radial direction of the spike arm. Panels E and F have the added global average of the antibody bound subvolumes from the single arm classification shown in dodger blue.
The comparison of our difference peaks with the 3-D reconstruction of the soluble Env spike in complex with PGT135, as well as its crystal structure, was more difficult. The Fab density in the PGT135 map is less well defined than that of PGT128, so its crystal structure in complex with a gp120 construct could not be readily placed with any confidence within its map. We therefore aligned the crystal structure of the PGT135-spike arm construct with the PGT128-construct and then aligned the two negative stain reconstructions to each other within this constraint. The combined PGT crystal structures and density maps could then be readily aligned to our unliganded Env spike density. When this is done, again the difference peaks for the six classes are closely positioned at the base of the PGT135 epitope site (Fig. 4D; see also Movie S6 in the supplemental material). Thus, these two independent reconstructions support the interpretation of our data, that the crown of the V3 loop, when in complex with 447-52D, is radially positioned and closer to the base of a flexible V3 loop rather than fixed near the spike apex, at least in the easily neutralized HIV-1 MN strain.
The general pattern that has emerged from the cryo-EM studies to date is that untriggered spikes have a closed configuration and CD4-liganded spikes have an open configuration (47, 48, 50). Variations on this theme include partial opening induced by binding to CD4bs MAbs (e.g., b12) and constitutive open configurations on some easily neutralizable forms (48, 64). The results reported here show that 447-52D binding also induces a conformational change in the spike toward the open conformation. The open conformation normally makes V3 available for coreceptor binding following CD4 ligation (20, 65). Because 447-52D binds to part of the coreceptor binding site the most likely mechanism of 447-52D neutralization would appear to be disruption or blockage of the coreceptor binding site. This need not be exclusive of other changes because V3 loop flexibility when antibody bound could alter the spike conformation such as altering or disordering the V1/V2 loop or other, more subtle ways that cannot be discerned at our resolution.
Conclusions.
The rearrangement and exposure of the V3 loop upon CD4 ligation followed by coreceptor binding is a key step in the viral infection cycle. The observation that MAb 447-52D Fab arms, which target the crown of the V3 loop, project radially rather than apically, as would have been predicted based on previous structural modeling, was unexpected. Either the crown is, in fact, localized to this position, is only exposed to this MAb when sampling this position, or reconfigures to this position from another upon 447-52D liganding. From our data, we can propose that 447-52D exerts its neutralizing effect by binding to the crown of the V3 loop and either locking it into a configuration incapable of binding the coreceptor or negatively altering other structural spike components that are necessary for efficient membrane fusion. The result indicates an interesting symmetry. CD4 binding induces a change to the open conformation exposing the V3 loop to coreceptor binding. Binding of 447-52D to an exposed V3 loop prevents reestablishment of the closed conformation, thereby locking the Env spike in the open conformation. This scenario does not rule out the possibility that this antibody may also exert its neutralizing effect during the extensive structural rearrangements that occur during the triggering and fusion process.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants 2R01 AI 055461 (to K.H.R. and K.A.T.), by P01 AI100151 (to Xiang-Peng Kong and Susan Zolla-Pazner), and by research funds from the Department of Veterans Affairs. Virus samples were generously supplied by the AIDS Vaccine Program (SAIC Frederick, NCI, Frederick, MD).
We thank Elena Chertova and Julian Bess, Jr., for production and biochemical analyses of the viruses; Zhong Huang for get_diffmap.py and advice on the use of the MMTSB tool set, SPARX, and Spider software packages; Guiqing Hu for advice on the one-arm classification; Hanspeter Winkler for help with the protomo software; and Susan Zolla-Pazner for supplying MAb 447-52D and for critically reviewing an earlier version of the manuscript.
Footnotes
Published ahead of print 13 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01596-14.
REFERENCES
- 1.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Doms RW, Moore JP. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9–F14. 10.1083/jcb.151.2.F9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354–2360. 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J. 2003. Public health: AIDS vaccine trial produces disappointment and confusion. Science 299:1290–1291. [DOI] [PubMed] [Google Scholar]
- 5.Lifson JD, Rossio JL, Piatak M, Jr, Bess J, Jr, Chertova E, Schneider DK, Coalter VJ, Poore B, Kiser RF, Imming RJ, Scarzello AJ, Henderson LE, Alvord WG, Hirsch VM, Benveniste RE, Arthur LO. 2004. Evaluation of the safety, immunogenicity, and protective efficacy of whole inactivated simian immunodeficiency virus (SIV) vaccines with conformationally and functionally intact envelope glycoproteins. AIDS Res. Hum. Retrovir. 20:772–787. 10.1089/0889222041524661. [DOI] [PubMed] [Google Scholar]
- 6.Sekaly RP. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 205:7–12. 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acharya P, Luongo TS, Matz J, Schmidt SD, Chuang GY, Georgiev I, Kessler P, Yang Y, Chames P, Martin L, Mascola JR, Kwong PD. 2012. Structural definition of a novel CD4-induced epitope that is targeted by a single-headed immunoglobulin to effect broad and potent HIV neutralization. Retrovirology 9:346. 10.1186/1742-4690-9-S2-P346. [DOI] [Google Scholar]
- 8.Liu L, Cimbro R, Lusso P, Berger EA. 2011. Intraprotomer masking of third variable loop (V3) epitopes by the first and second variable loops (V1V2) within the native HIV-1 envelope glycoprotein trimer. Proc. Natl. Acad. Sci. U. S. A. 108:20148–20153. 10.1073/pnas.1104840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusert P, Krarup A, Magnus C, Brandenberg OF, Weber J, Ehlert AK, Regoes RR, Gunthard HF, Trkola A. 2011. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J. Exp. Med. 208:1419–1433. 10.1084/jem.20110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892–10905. 10.1128/JVI.75.22.10892-v10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 15.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modrow S, Hahn BH, Shaw GM, Gallo RC, Wong-Staal F, Wolf H. 1987. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J. Virol. 61:570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoxie JA. 2010. Toward an antibody-based HIV-1 vaccine. Annu. Rev. Med. 61:135–152. 10.1146/annurev.med.60.042507.164323. [DOI] [PubMed] [Google Scholar]
- 19.Laakso MM, Lee FH, Haggarty B, Agrawal C, Nolan KM, Biscone M, Romano J, Jordan AP, Leslie GJ, Meissner EG, Su L, Hoxie JA, Doms RW. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3:e117. 10.1371/journal.ppat.0030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zolla-Pazner S, Cardozo T. 2010. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 10:527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaRosa GJ, Davide JP, Weinhold K, Waterbury JA, Profy AT, Lewis JA, Langlois AJ, Dreesman GR, Boswell RN, Shadduck P. 1990. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science 249:932–935. 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- 22.Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. 2004. The v3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394–2404. 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, Zolla-Pazner S. 2010. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS One 5:e10254. 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweighardt B, Liu Y, Huang W, Chappey C, Lie YS, Petropoulos CJ, Wrin T. 2007. Development of an HIV-1 reference panel of subtype B envelope clones isolated from the plasma of recently infected individuals. J. Acquir. Immune Defic. Syndr. 46:1–11. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125. 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 79:10103–10107. 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808–9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, Morris L, Salomon A, Pinter A, Hoxie JA, Hahn BH, Kwong PD, Shaw GM. 2009. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J. Virol. 83:1240–1259. 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson WE, Morgan J, Reitter J, Puffer BA, Czajak S, Doms RW, Desrosiers RC. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075–2086. 10.1128/jvi.76.5.2075-2086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435–3443. 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J. Virol. 79:780–790. 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pantophlet R, Burton DR. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739–769. 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 33.Saunders CJ, McCaffrey RA, Zharkikh I, Kraft Z, Malenbaum SE, Burke B, Cheng-Mayer C, Stamatatos L. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069–9080. 10.1128/JVI.79.14.9069-9080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatatos L, Cheng-Mayer C. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840–7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lusso P, Earl PL, Sironi F, Santoro F, Ripamonti C, Scarlatti G, Longhi R, Berger EA, Burastero SE. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J. Virol. 79:6957–6968. 10.1128/JVI.79.11.6957-6968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384–9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035–9045. 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841. 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Lu M, Poon BK, Wang Q, Ma J. 2009. Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement. Acta Crystallogr. D Biol. Crystallogr. 65:339–347. 10.1107/S0907444909003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166–1171. 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852. 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 46.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2:e83. 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249. 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu G, Liu J, Taylor KA, Roux KH. 2011. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J. Virol. 85:2741–2750. 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J. Virol. 85:12114–12123. 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramanian S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445. 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chertova E, Crise BJ, Morcock DR, Bess JW, Jr, Henderson LE, Lifson JD. 2003. Sites, mechanism of action and lack of reversibility of primate lentivirus inactivation by preferential covalent modification of virion internal proteins. Curr. Mol. Med. 3:265–272. 10.2174/1566524033479889. [DOI] [PubMed] [Google Scholar]
- 53.Rossio JL, Esser MT, Suryanarayana K, Schneider DK, Bess JW, Jr, Vasquez GM, Wiltrout TA, Chertova E, Grimes MK, Sattentau Q, Arthur LO, Henderson LE, Lifson JD. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winkler H, Taylor KA. 2006. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 106:240–254. 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 55.Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. 2009. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J. Struct. Biol. 165:64–77. 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkler H, Taylor KA. 1999. Multivariate statistical analysis of three-dimensional cross-bridge motifs in insect flight muscle. Ultramicroscopy 77:141–152. 10.1016/S0304-3991(99)00035-2. [DOI] [Google Scholar]
- 57.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46. 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. 2004. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure 12:193–204. 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Tama F, Miyashita O, Brooks CL., III 2004. Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryo-EM. J. Struct. Biol. 147:315–326. 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 60.Feig M, Karanicolas J, Brooks CL., III 2004. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graph Model 22:377–395. [DOI] [PubMed] [Google Scholar]
- 61.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Roux KH, Taylor KA. 2007. AIDS virus envelope spike structure. Curr. Opin. Struct. Biol. 17:244–252. 10.1016/j.sbi.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 19:893–899. 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8:e1002797. 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cormier EG, Dragic T. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953–8957. 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spear GT, Takefman DM, Sharpe S, Ghassemi M, Zolla-Pazner S. 1994. Antibodies to the HIV-1 V3 loop in serum from infected persons contribute a major proportion of immune effector functions including complement activation, antibody binding, and neutralization. Virology 204:609–615. 10.1006/viro.1994.1575. [DOI] [PubMed] [Google Scholar]
- 67.Javaherian K, Langlois AJ, McDanal C, Ross KL, Eckler LI, Jellis CL, Profy AT, Rusche JR, Bolognesi DP, Putney SD. 1989. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc. Natl. Acad. Sci. U. S. A. 86:6768–6772. 10.1073/pnas.86.17.6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swetnam J, Shmelkov E, Zolla-Pazner S, Cardozo T. 2010. Comparative magnitude of cross-strain conservation of HIV variable loop neutralization epitopes. PLoS One 5:e15994. 10.1371/journal.pone.0015994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. U. S. A. 109:5663–5668. 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 84:3147–3161. 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538–7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635–643. [PubMed] [Google Scholar]
- 73.Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232–13252. 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Almond D, Kimura T, Kong X, Swetnam J, Zolla-Pazner S, Cardozo T. 2010. Structural conservation predominates over sequence variability in the crown of HIV type 1's V3 loop. AIDS Res. Hum. Retrovir. 26:717–723. 10.1089/aid.2009.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keller PM, Arnold BA, Shaw AR, Tolman RL, Van Middlesworth F, Bondy S, Rusiecki VK, Koenig S, Zolla-Pazner S, Conard P. 1993. Identification of HIV vaccine candidate peptides by screening random phage epitope libraries. Virology 193:709–716. 10.1006/viro.1993.1179. [DOI] [PubMed] [Google Scholar]
- 76.Rees WA, Keller RW, Vesenka JP, Yang G, Bustamante C. 1993. Evidence of DNA bending in transcription complexes imaged by scanning force microscopy. Science 260:1646–1649. 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- 77.Burke V, Williams C, Sukumaran M, Kim SS, Li H, Wang XH, Gorny MK, Zolla-Pazner S, Kong XP. 2009. Structural basis of the cross-reactivity of genetically related human anti-HIV-1 MAbs: implications for design of V3-based immunogens. Structure 17:1538–1546. 10.1016/j.str.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conley AJ, Conard P, Bondy S, Dolan CA, Hannah J, Leanza WJ, Marburg S, Rivetna M, Rusiecki VK, Sugg EE. 1994. Immunogenicity of synthetic HIV-1 gp120 V3-loop peptide-conjugate immunogens. Vaccine 12:445–451. [DOI] [PubMed] [Google Scholar]
- 79.Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong XP. 2010. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol. 17:955–961. 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- 80.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452. 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.VanCott TC, Bethke FR, Polonis VR, Gorny MK, Zolla-Pazner S, Redfield RR, Birx DL. 1994. Dissociation rate of antibody-gp120 binding interactions is predictive of V3-mediated neutralization of HIV-1. J. Immunol. 153:449–459. [PubMed] [Google Scholar]
- 82.Zhu P, Chertova E, Bess J, Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. U. S. A. 100:15812–15817. 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu P, Winkler H, Chertova E, Taylor KA, Roux KH. 2008. Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. PLoS Pathog. 4:e1000203. 10.1371/journal.ppat.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528. 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 85.Chertova E, Bess JW, Jr, Crise BJ, Sowder IR, Schaden TM, Hilburn JM, Hoxie JA, Benveniste RE, Lifson JD, Henderson LE, Arthur LO. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315–5325. 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klein JS, Bjorkman PJ. 2010. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6:e1000908. 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kwong PD, Wilson IA. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 10:573–578. 10.1038/ni.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dhillon AK, Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. 2008. Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set. Acta Crystallogr. D Biol. Crystallogr. D. 64:792–802. 10.1107/S0907444908013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 20:796–803. 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


