FIG 5.
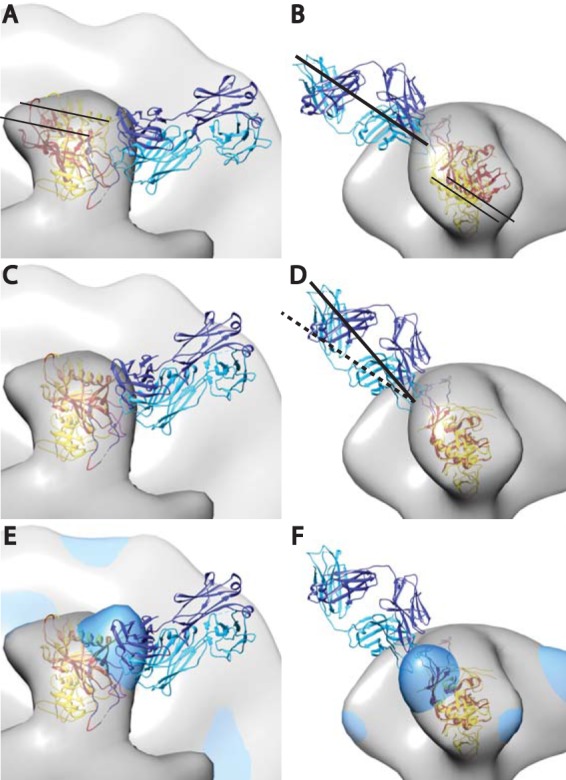
Crystal structure of PGT128 Fab in complex with an engineered glycosylated gp120 outer domain with a miniV3 loop (eODmV3), PDB 3TYG (1), aligned onto the gp120 spike pseudoatomic model from the present study. The 3TYG crystal structure is shown in orange-red, and the gp120 pseudoatomic model from the present work shown in yellow. The PGT128 Fab is shown in blue and cyan. The symmetrized average Env spike from the present study is provided as a point of reference. Panels A and B show the crystal structure as it would be positioned when fitted in the PGT128 image reconstruction (emd-1970), followed by alignment of emd-1970 to our control spike reconstruction. Panels C and D are after alignment on the gp120 atomic model used in the present study. The alignment was done manually using CHIMERA. The major movement requires a small outward radial movement and a small downward axial movement as indicated by the narrow black lines. There is also an ∼15° clockwise rotation along an axis roughly perpendicular to the radial direction of the spike arm. Panels E and F have the added global average of the antibody bound subvolumes from the single arm classification shown in dodger blue.
