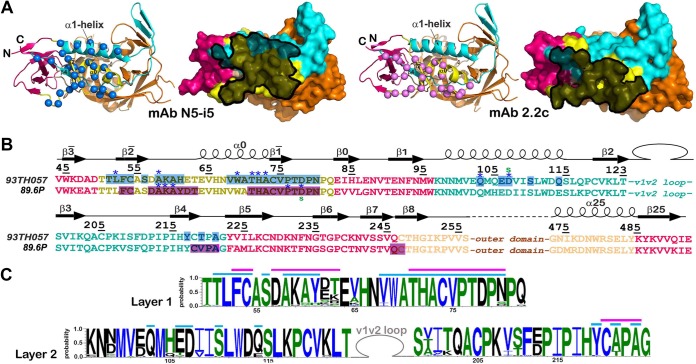
FIG 3.
N5-i5 and 2.2c epitopes. (A) N5-i5 and 2.2c epitope footprints on monomeric gp120. The Cα atoms of the gp120 residues involved in N5-i5 and 2.2c binding are represented by blue and pink balls, respectively, and displayed over the ribbon diagram of the gp120 inner domain. Antibodies' contact surfaces displayed over the gp120 molecular surface are shown in black (right). The “layered” architecture of the gp120 inner domain is shown, with the 7-stranded β sandwich in magenta, layer 1 in yellow, layer 2 in cyan, and layer 3 in light orange. The outer domain is shown in orange. (B) Mapping of the N5-i5 and 2.2c contact residues on the gp120 primary sequence of the gp120 inner domain of the isolates used in structural studies. The topology diagrams depicting a distribution of secondary structure elements as calculated with DSSP (66) is shown above the gp120 sequences. The gp120 residues involved in N5-i5 and 2.2c binding are highlighted in blue and pink, respectively. Residues contributing to the binding through H bonds and salt bridges are indicated by blue asterisks and green lowercase letters above and below the 93TH057 and 89.6P sequences, respectively. The gp120 layers are colored as in panel A. (C) Sequence conservation of N5-i5 and 2.2c epitopes. The height of the residue at each position is proportional to its frequency of distribution among the HIV-1 isolates, as deposited in the Los Alamos database (all clades are included). Residues are colored according to hydrophobicity: black, hydrophilic; green, neutral; blue, hydrophobic. Residues forming the N5-i5 and 2.2c epitope are indicated by blue and pink lines above the sequence, respectively.