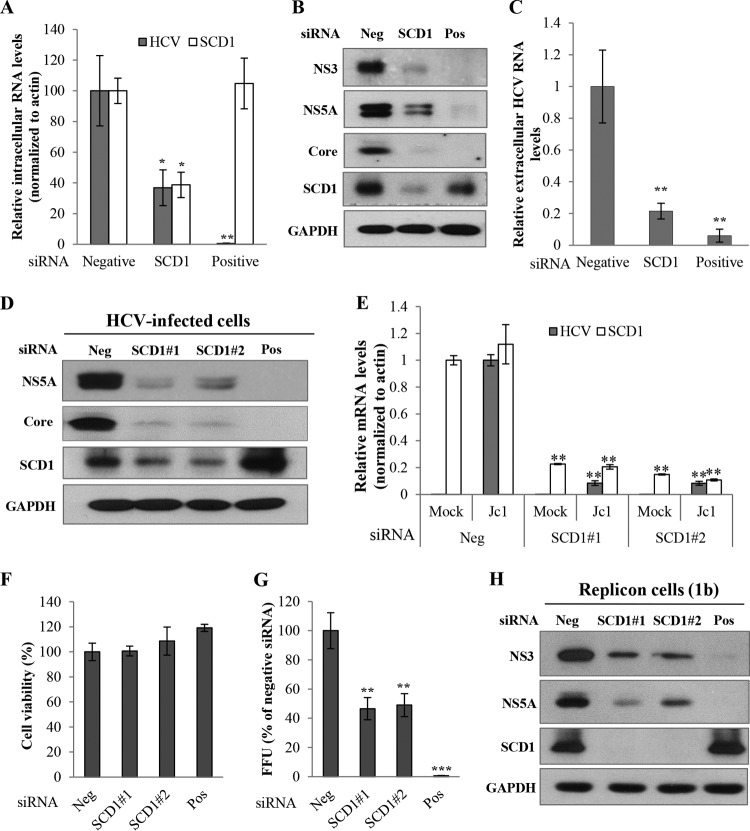
FIG 1.
SCD1 is required for HCV replication. (A) Huh7.5 cells were transfected with 10 nM siRNA pool targeting four different sites of SCD1 mRNA for 48 h and then infected with Jc1 for 4 h. At 2 days postinfection, intracellular RNA levels of both HCV and SCD1 were quantified by quantitative PCR. (B) Total cell lysates were immunoblotted with the indicated antibodies. (C) Extracellular RNAs isolated from the culture supernatant were quantified by qPCR. Negative, scrambled siRNA; positive, HCV-specific siRNA. (D) Huh7.5 cells were transfected with the indicated siRNAs for 48 h and then infected with Jc1 for 4 h. At 2 days postinfection, total cell lysates were immunoblotted with the indicated antibodies. Suffixes #1 and #2 refer to different SCD1 siRNA sequences. (E) Huh7.5 cells were treated as described for panel D, and intracellular RNAs were quantified by quantitative PCR. (F) Cell viability was determined by MTT assay. (G) Huh7.5 cells were transfected with the indicated siRNAs for 48 h and then infected with Jc1 for 4 h. At 2 days postinfection, culture supernatant was harvested and was used to infect naive Huh7.5 cells. HCV infectivity was determined by FFU/ml. Experiments were performed in triplicate. Error bars indicate the standard deviations of the means. (H) Huh7 cells harboring HCV replicon derived from genotype 1b were transfected with the indicated siRNAs. At 3 days after transfection, total cell lysates were immunoblotted with the indicated antibodies. Data represent averages from at least three independent experiments for panels A, C, E, F, and G. The asterisks indicate significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) from the value for the negative control.